Journal:IUCrJ:S2052252521005340
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
<scene name='88/884898/Cv/25'>Anti-HIV antibody RoAb13</scene> targets a short N-terminal region of the protein CCR5, which is the main entry receptor of HIV into the human organism. Blocking that receptor would therefore prevent HIV infection and replication. We had earlier reported the structure of the antibody alone by X-ray crystallography (Chain ''et al.'' 2015<ref name="Chain">PMID:26030924</ref>), but the structure of the antibody complexed to the part of CCR5 to which it binds (its epitope) had remained elusive. That structure is important for designing efficient vaccines based on short synthetic immunogenic peptides that mimic the CCR5 antibody-binding region. | <scene name='88/884898/Cv/25'>Anti-HIV antibody RoAb13</scene> targets a short N-terminal region of the protein CCR5, which is the main entry receptor of HIV into the human organism. Blocking that receptor would therefore prevent HIV infection and replication. We had earlier reported the structure of the antibody alone by X-ray crystallography (Chain ''et al.'' 2015<ref name="Chain">PMID:26030924</ref>), but the structure of the antibody complexed to the part of CCR5 to which it binds (its epitope) had remained elusive. That structure is important for designing efficient vaccines based on short synthetic immunogenic peptides that mimic the CCR5 antibody-binding region. | ||
| + | |||
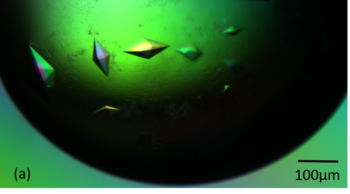
| + | [[Image:Figure_1aaa.png|thumb|350px|left|'''Figure 1a''' Crystals of RoAb13 complexed with the 31 peptide. X-ray crystallography.]] | ||
| + | |||
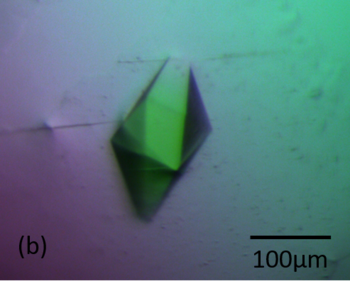
| + | [[Image:Figure_1bbb.png|thumb|350px|left|'''Figure 1b''' Crystals of RoAb13 complexed with the PIYDIN peptide]] | ||
After a long search for co-crystallisation conditions involving both the whole N-terminal region of CCR5 and the minimally required binding region to its antibody (the ‘core peptide’), and the analysis and comparison of X-ray crystallographic electron density maps obtained from several crystals, we have finally located the core peptide of the CCR5 receptor bound to RoAb13. It binds at the hypervariable region ‘CDR3’ of the antibody’s light chain, an expected antigen-binding site. In spite of the fact that the best attainable resolution is not particularly high at 3 Å, we have been able to identify the interacting residues between antibody and peptide: | After a long search for co-crystallisation conditions involving both the whole N-terminal region of CCR5 and the minimally required binding region to its antibody (the ‘core peptide’), and the analysis and comparison of X-ray crystallographic electron density maps obtained from several crystals, we have finally located the core peptide of the CCR5 receptor bound to RoAb13. It binds at the hypervariable region ‘CDR3’ of the antibody’s light chain, an expected antigen-binding site. In spite of the fact that the best attainable resolution is not particularly high at 3 Å, we have been able to identify the interacting residues between antibody and peptide: | ||
| Line 14: | Line 18: | ||
Furthermore, the core peptide was found to bind in a way that can accommodate the full length of the CCR5 N-terminus. | Furthermore, the core peptide was found to bind in a way that can accommodate the full length of the CCR5 N-terminus. | ||
The structural insights thus may inform the design of better peptide analogues for use as immunogens in vivo. These analogues may ultimately provide the basis for active immunization vaccines to stimulate an antibody response to native CCR5 which will thwart HIV infection. | The structural insights thus may inform the design of better peptide analogues for use as immunogens in vivo. These analogues may ultimately provide the basis for active immunization vaccines to stimulate an antibody response to native CCR5 which will thwart HIV infection. | ||
| - | |||
| - | [[Image:Figure_1aaa.png|thumb|350px|left|'''Figure 1a''' Crystals of RoAb13 complexed with the 31 peptide. X-ray crystallography.]] | ||
| - | |||
| - | [[Image:Figure_1bbb.png|thumb|350px|left|'''Figure 1b''' Crystals of RoAb13 complexed with the PIYDIN peptide]] | ||
'''The PIYDIN of the 31 peptide study (Figure 2a):''' | '''The PIYDIN of the 31 peptide study (Figure 2a):''' | ||
Revision as of 10:47, 14 June 2021
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.