Journal:IUCrJ:S2052252521008125
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
We follow the increase of the CEF occupancy in the catalytic cleft of BlaC from 5 ms to 50 ms after mixing with CEF by XFEL based MISC experiments. From the increasing occupancy values we are able to estimate an effective diffusion coefficient Deff of CEF in the BlaC microcrystals. Diffusion is roughly an order of magnitude slower in the crystals than that in water. By using the Deff we are now able to calculate CEF occupancies anywhere in the microcrystals and at any time. Since the BlaC-CEF complex formation triggers the enzymatic cycle, the speed of its formation determines the time-resolution of the MISC method. Our results show that the time-resolution is on the order of 5 ms for our microcrystals. The structures of any intermediates whose lifetimes are longer can therefore be characterized. Once faster intermediates are concerned, smaller crystals must be used that support a faster buildup of the enzyme substrate complex. | We follow the increase of the CEF occupancy in the catalytic cleft of BlaC from 5 ms to 50 ms after mixing with CEF by XFEL based MISC experiments. From the increasing occupancy values we are able to estimate an effective diffusion coefficient Deff of CEF in the BlaC microcrystals. Diffusion is roughly an order of magnitude slower in the crystals than that in water. By using the Deff we are now able to calculate CEF occupancies anywhere in the microcrystals and at any time. Since the BlaC-CEF complex formation triggers the enzymatic cycle, the speed of its formation determines the time-resolution of the MISC method. Our results show that the time-resolution is on the order of 5 ms for our microcrystals. The structures of any intermediates whose lifetimes are longer can therefore be characterized. Once faster intermediates are concerned, smaller crystals must be used that support a faster buildup of the enzyme substrate complex. | ||
| + | |||
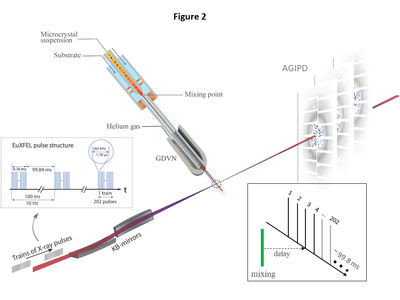
| + | [[Image:Figure2Schmidt.png|left|400px|thumb|Figure 2. Experimental setup at the European XFEL. BlaC microcrystals are mixed with substrate and | ||
| + | injected into the X-ray interaction region (dotted circle) after a delay determined by the distance between | ||
| + | the mixing region and the X-rays, the capillary width, and the flow rate. Diffusion of substrate into the | ||
| + | crystals occurs during this time. The mixture is probed by trains of X-ray pulses. The trains repeat 10 | ||
| + | times per second. Pulses within the trains repeat with 564 kHz, hence the pulses are spaced by 1.78 μs. | ||
| + | 202 pulses were in each train for this experiment. The AGIPD collects the diffraction patterns and reads | ||
| + | them out for further analysis. Inset, data collection: With a once selected injector geometry and flow | ||
| + | rate, the delay is fixed by the distance of the mixing region from the X-ray interaction region. All pulses | ||
| + | in all trains (here pulse #3) probe the same time delay. The EuXFEL pulse structure is most efficiently | ||
| + | used.]] | ||
| + | {{Clear}} | ||
In addition to the CEF binding we observe the reaction of BlaC with an inhibitor sulbactam (SUB) at 66 ms. SUB reacts to a so-called trans enamine in subunits B and D of the BlaC already after 66 ms. However, SUB stays intact in subunits A and C. As the reaction proceeds to the trans-enamine also in A and C, the structures of the weakly bound SUBs in these subunits at 66 ms represent an interesting intermediate that would not have been detected in static structures. | In addition to the CEF binding we observe the reaction of BlaC with an inhibitor sulbactam (SUB) at 66 ms. SUB reacts to a so-called trans enamine in subunits B and D of the BlaC already after 66 ms. However, SUB stays intact in subunits A and C. As the reaction proceeds to the trans-enamine also in A and C, the structures of the weakly bound SUBs in these subunits at 66 ms represent an interesting intermediate that would not have been detected in static structures. | ||
Revision as of 09:44, 8 August 2021
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.