Aminoacyl tRNA Synthetase
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
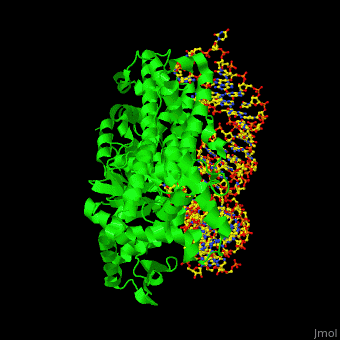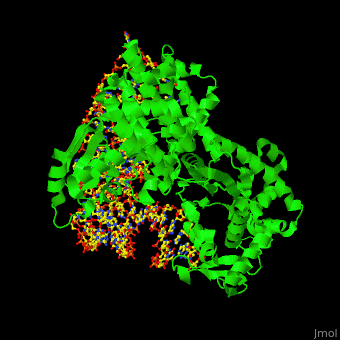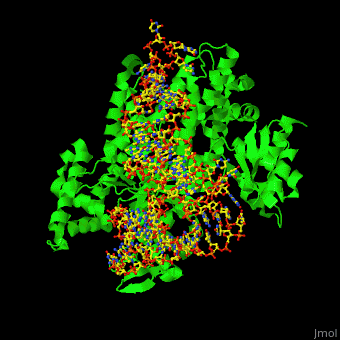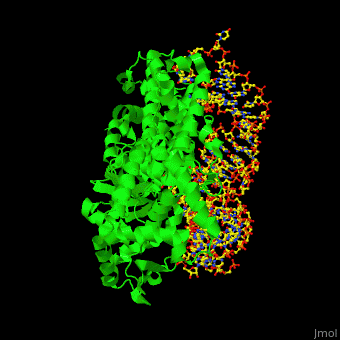
The <scene name='44/444597/Protein_surface_interaction/2'>glutaminyl-tRNA synthetase</scene> (GlnRS) interacts with both <scene name='44/444597/Acceptor_and_anticodon_interac/1'>the anticodon loop and the acceptor stem</scene>. Genetic and biochemical data indicate that GlnRS interacts with all three bases of the <scene name='44/444597/Anticodon_loop/3'>anticodon loop</scene>, which are unstacked and splay outward so they can bind in separate recognition pockets of GlnRS. The 3' end of tRNAgln plunges deeply into a protein pocket that also binds the enzyme's <scene name='44/444597/Atp_binding_site/1'>ATP</scene> and glutamine substrates. | The <scene name='44/444597/Protein_surface_interaction/2'>glutaminyl-tRNA synthetase</scene> (GlnRS) interacts with both <scene name='44/444597/Acceptor_and_anticodon_interac/1'>the anticodon loop and the acceptor stem</scene>. Genetic and biochemical data indicate that GlnRS interacts with all three bases of the <scene name='44/444597/Anticodon_loop/3'>anticodon loop</scene>, which are unstacked and splay outward so they can bind in separate recognition pockets of GlnRS. The 3' end of tRNAgln plunges deeply into a protein pocket that also binds the enzyme's <scene name='44/444597/Atp_binding_site/1'>ATP</scene> and glutamine substrates. | ||
| + | |||
| + | One interesting question is how tRNA synthetases are able to discern that the correct amino acid is loaded. The structure for aspartate is very similar to glutamine, differing only by an amino group and one CH2 group. However, the aminoacyl tRNA synthetases interact with the tRNA in completely different fashions. Asp-tRNA synthetase acts as a <scene name='44/444597/Asp_trna_synthetase/1'>dimer</scene>, and interacts with the opposite face of the tRNA, the <scene name='44/444597/Major_groove_binding/1'>major groove</scene> side. The anticodon arm of tRNA Asp is bent by as much as 20 A toward the inside of the L relative to that in the X-ray structure of uncomplexed tRNA Asp, and its anticodon bases are unstacked. The hinge point for the bend is a <scene name='44/444597/U40_g30_h_bond/1'>G30 • U40</scene> base pair in the anticodon stem (nearly all other species of tRNA contain a Watson-Crick base pair at that point) | ||
</StructureSection> | </StructureSection> | ||
Revision as of 03:49, 23 September 2021
| |||||||||||
References
- ↑ Cavarelli J, Moras D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J. 1993 Jan;7(1):79-86. PMID:8422978
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Ann Taylor