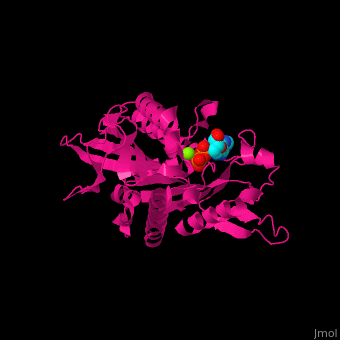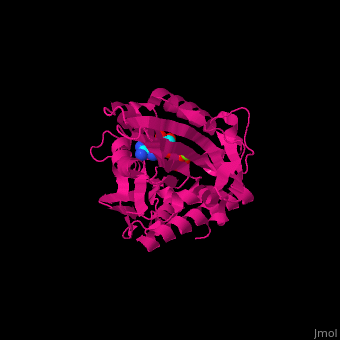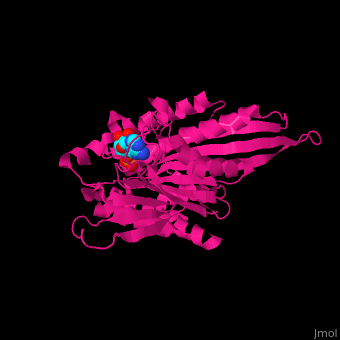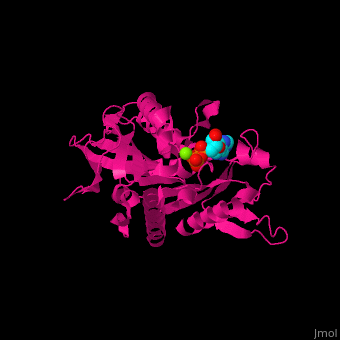Kinesin
From Proteopedia
(Difference between revisions)
| (62 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='350' side='right' scene='41/410296/Cv/1' caption='KIF1A Motor Domain complex with ADP and Mg+2 ion (green), [[2zfi]]'> | |
| - | + | == Function == | |
| - | + | ||
| - | + | [[Kinesin|Kinesins]] are eukaryotic motor proteins which move along microtubules<ref>PMID:19773780</ref>. Kinesin (KIF) is a dimer consisting of 2 heavy chains and two light chains. The heavy chain contains the N-terminal globular motor domain (MD) residues 1-375, responsible for the motor activity of kinesin, a central flexible neck linker (FNL) coiled-coil stalk which intertwines to form the dimer and a small globular C-terminal domain which interacts with other proteins like the kinesin light chain. The light chain (KLC) forms the tail region. The KLC contains a cargo binding domain which is called TPR (Tetratricopeptide repeat). The KIFs are named by their gene number. KIF are organized into 14 families named kinesin-1 to kinesin-14. KIF contains a forkhead-associated domain (FHA) which is involved in phosphopeptide recognition.<br /> | |
| - | Kinesins are eukaryotic motor proteins which move along microtubules. Kinesin (KIF) is a dimer consisting of 2 heavy chains and two light chains. The heavy chain contains the globular motor domain (MD), flexible neck linker (FNL) | + | *'''KIF1A''' transports organelles along axonal microtubules.<br /> |
| + | *'''KIF1C''' and '''KIF2''' are plus-ended directed microtubule motors.<br /> | ||
| + | *'''KIF2C''', '''KIF22''' and '''KIF3B''' are plus-ended directed microtubule motors in mitotic cells<br /> | ||
| + | *'''KIF13B''' transports VEGFR2 from the Golgi to the endothelial cell surface.<br /> | ||
| + | *'''KIF16B''' is plus-ended directed microtubule motor involved in endosome transport and receptor recycling.<br /> | ||
| + | *'''KIFC1''' transports DNA molecules along cytoskeleton filaments.<br /> | ||
| + | *'''Kar3''' is kinesin protein in yeast.<br /> | ||
| + | *'''Eg5''' or '''KIF11''' is a kinesin (See [[Kinesin-5]]) which participates in mitosis.<br /> | ||
| + | *'''NOD''' is a ''Drosophila'' chromosome-associated kinesin. | ||
| + | See also [[CAP-Gly domain]]. | ||
| - | == | + | == Disease == |
| + | Mutations in KIF5A are involved in hereditary spastic paraplegia<ref>PMID:18203753</ref>. Mutation in KIF1B is the cause of Charcot-Marie-Tooth disease <ref>PMID:14559185</ref>. Mutations in KIF22 cause spondyloepimetaphyseal dysplasia<ref>PMID:23665482</ref>. | ||
| - | === | + | == Structural highlights == |
| + | <scene name='41/410296/Cv/10'>Active site</scene>. Water molecules shown as red spheres. Residue <scene name='41/410296/Cv/11'>Arg216 is the key residue in KIF for the chemical cycling of ATPase and for the mechanical cycling</scene>. Arg216 pivots to enable <scene name='41/410296/Cv/12'>Mg-ADP</scene> release or the phosphate release. <scene name='41/410296/Cv/13'>Arg216 forms a latch</scene> in the KIF 'closed-state' before the Mg-ADP release. Binding of β-tubulin to KIF releases the latch, enabling the KIF conformation change and detaching KIF from the microtubule and enabling the next movement cycle<ref>PMID:18806800</ref>. | ||
| - | + | <scene name='41/410296/Cv/14'>Mg coordination site</scene>. | |
| - | + | == 3D Structures of Kinesin == | |
| - | + | [[Kinesin 3D Structures]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | </StructureSection> | ||
| - | == | + | == References == |
| - | + | <references/> | |
| - | [[ | + | [[Category:Topic Page]] |
| - | + | ||
Current revision
| |||||||||||
References
- ↑ Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009 Oct;10(10):682-96. doi: 10.1038/nrm2774. PMID:19773780 doi:http://dx.doi.org/10.1038/nrm2774
- ↑ Ebbing B, Mann K, Starosta A, Jaud J, Schols L, Schule R, Woehlke G. Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum Mol Genet. 2008 May 1;17(9):1245-52. doi: 10.1093/hmg/ddn014. Epub 2008 Jan, 18. PMID:18203753 doi:http://dx.doi.org/10.1093/hmg/ddn014
- ↑ Hirokawa N, Takemura R. Biochemical and molecular characterization of diseases linked to motor proteins. Trends Biochem Sci. 2003 Oct;28(10):558-65. PMID:14559185 doi:http://dx.doi.org/10.1016/j.tibs.2003.08.006
- ↑ Grosch M, Gruner B, Spranger S, Stutz AM, Rausch T, Korbel JO, Seelow D, Nurnberg P, Sticht H, Lausch E, Zabel B, Winterpacht A, Tagariello A. Identification of a Ninein (NIN) mutation in a family with spondyloepimetaphyseal dysplasia with joint laxity (leptodactylic type)-like phenotype. Matrix Biol. 2013 Oct-Nov;32(7-8):387-92. doi: 10.1016/j.matbio.2013.05.001. Epub, 2013 May 9. PMID:23665482 doi:http://dx.doi.org/10.1016/j.matbio.2013.05.001
- ↑ Nitta R, Okada Y, Hirokawa N. Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat Struct Mol Biol. 2008 Oct;15(10):1067-75. Epub 2008 Sep 21. PMID:18806800 doi:10.1038/nsmb.1487
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, Jaime Prilusky