BamHI
From Proteopedia
(Difference between revisions)
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | <StructureSection load='1bhm' size='350' side='right' scene='' caption='BamHI dimer complex with DNA (PDB code [[1bhm]])'> | ||
[[Image:1bam.png|left|200px]] | [[Image:1bam.png|left|200px]] | ||
| - | + | __TOC__ | |
| - | + | ||
| - | + | ||
===Overview=== | ===Overview=== | ||
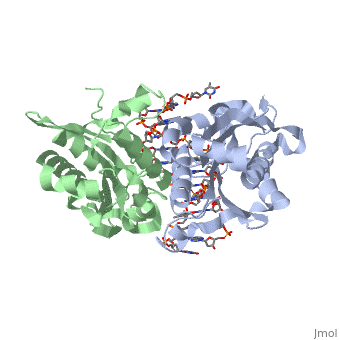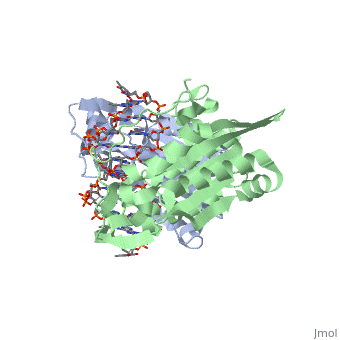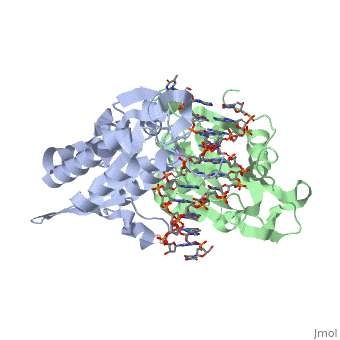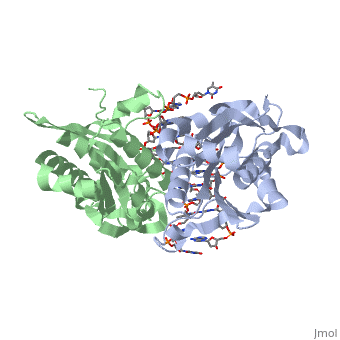
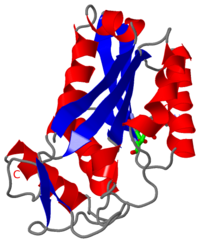
| - | <scene name='BamHI/1bhm/3'>BamHI</scene> is a type II restriction enzyme derived from ''Bacillus amyloliquefaciens''. Like all Type II restriction endonucleases, it is a <scene name=' | + | <scene name='BamHI/1bhm/3'>BamHI</scene> is a type II restriction enzyme derived from ''Bacillus amyloliquefaciens''. Like all Type II restriction endonucleases, it is a <scene name='47/472560/Dimer/1'>dimer</scene> and the recognition site is palindromic and 6 bases in length. It recognizes the DNA sequence of G’GATCC and leaves an overhang of GATC which is compatible with many other enzymes.<ref name="Viadiu">Viadiu H, Aggarwal AK, Structure of BamHI bound to nonspecific DNA: a model for DNA sliding., ''Mol Cell.'' 2000 May;5(5):889-95. Print.</ref> The ''Bam''HI-DNA complex is a sequence-specific endonucleases-DNA complex. In the complex all hydrogen bonding occurs in the <scene name='47/472560/Major_groove/1'>major groove</scene> of the recognition site,either through direct or ''water-mediated'' hydrogen bonds with the protein, namely on oxygens and nitrogens in the DNA within 3.5 angstroms of the protein. No other DNA sequence could support this degree of complementarity with ''Bam''HI.<ref name="Voet">Voet, Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. p. 578-579. Print.</ref> There are five crystal structures of ''Bam''HI in the Protein Data Bank. These include ''Bam''HI bound to a non-specific DNA, ''Bam''HI complex with DNA and calcium ions (pre-reactive complex), BamHI complex with DNA and manganese ions (post-reactive complex), ''Bam''HI complex with DNA, and ''Bam''HI phased at 1.95 angstroms resolution by MAD analysis.<ref name="pdbe">PDBe., ''European Molecular Biology Laboratory.'' 2011. Webpage.</ref> See also [[Endonuclease]]. |
| - | + | ||
| + | See also [[BamHI (Hebrew)]]. | ||
| + | |||
==Structure== | ==Structure== | ||
<scene name='BamHI/1bhm/8'>BamHI</scene> consists of 6 <scene name='BamHI/1bhm/10'>alpha helices</scene> and 6 <scene name='BamHI/1bhm/9'>beta sheets</scene>, surprisingly the protein sequence lacks similarity and has no recurring structural motifs analogous to the helix-turn-helix or the zinc finger of transcription factors.<ref name="pdbe">PDBe., ''European Molecular Biology Laboratory.'' 2011. Webpage.</ref> The beta sheet form a curved plane with only two sheets forming <scene name='BamHI/1bhm/11'>hydrogen bonds</scene> with specific DNA. BamHI is a dimer of 213 amino acid subunits. The structure of the protein allows <scene name='BamHI/1bhm/13'>hydrogen bonding</scene> to occur in the manner seen in this scene. | <scene name='BamHI/1bhm/8'>BamHI</scene> consists of 6 <scene name='BamHI/1bhm/10'>alpha helices</scene> and 6 <scene name='BamHI/1bhm/9'>beta sheets</scene>, surprisingly the protein sequence lacks similarity and has no recurring structural motifs analogous to the helix-turn-helix or the zinc finger of transcription factors.<ref name="pdbe">PDBe., ''European Molecular Biology Laboratory.'' 2011. Webpage.</ref> The beta sheet form a curved plane with only two sheets forming <scene name='BamHI/1bhm/11'>hydrogen bonds</scene> with specific DNA. BamHI is a dimer of 213 amino acid subunits. The structure of the protein allows <scene name='BamHI/1bhm/13'>hydrogen bonding</scene> to occur in the manner seen in this scene. | ||
==Active Sites and Catalytic Mechanism to Specific DNA== | ==Active Sites and Catalytic Mechanism to Specific DNA== | ||
| - | Type II restriction enzymes require only Mg2+ as a cofactor to catalyze the hydrolysis of DNA phosphodiesters, leaving free 5’ phosphate and 3’ hydroxyl groups. The reaction is considered to proceed by an inline displacement of the 3’ leaving group, in which an activated water molecule acts as the attacking nucleophile. The <scene name=' | + | Type II restriction enzymes require only Mg2+ as a cofactor to catalyze the hydrolysis of DNA phosphodiesters, leaving free 5’ phosphate and 3’ hydroxyl groups. The reaction is considered to proceed by an inline displacement of the 3’ leaving group, in which an activated water molecule acts as the attacking nucleophile. The <scene name='47/472560/Active_site/2'>active site</scene> for ''Bam''HI are residues Asp94, Glu111, and Glu113. These can be spatially aligned with residues in EcoRI, EcoRV and PvuII. Several mechanisms have been proposed for the way these enzymes might activate a water molecule for nucleophilic attack. It is suggested there is a general base mechanism in which the acidic residues Asp94 and Glu111 coordinate Mg2+ at the active site while Glu113 acts as a general base to deprotonate the attacking water molecule. A separate mechanism has been suggested in which one of the <scene name='47/472560/Active_site_water/1'>three water molecules</scene> located centrally at the active site within hydrogen-bonding distances from the carboxylate groups of Glu111 and Glu113 acts as a nucleophile and attacks the phosphate group of DNA.<ref name="Newman">Newman, Matthew, Structure of ''Bam''HI Endonuclease Bound to DNA: Partial Folding and Unfolding on DNA Binding., ''Science.'' 1995 Aug;4(269):656-63. Print.</ref> |
==Active Sites and Catalytic Mechanism for Binding to Nonspecific DNA== | ==Active Sites and Catalytic Mechanism for Binding to Nonspecific DNA== | ||
| Line 18: | Line 19: | ||
==3D structures of BamHI== | ==3D structures of BamHI== | ||
| - | [[1bhm]], [[3bam]], [[2bam]], [[1esg]] – | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} |
| + | |||
| + | [[1bhm]], [[3bam]], [[2bam]], [[1esg]] – BaBAM + DNA – ''Bacillus amyloliquefaciens''<br /> | ||
| + | [[1bam]] – BaBAM<br /> | ||
== References == | == References == | ||
Current revision
| |||||||||||
3D structures of BamHI
Updated on 17-January-2022
1bhm, 3bam, 2bam, 1esg – BaBAM + DNA – Bacillus amyloliquefaciens
1bam – BaBAM
References
- ↑ 1.0 1.1 Viadiu H, Aggarwal AK, Structure of BamHI bound to nonspecific DNA: a model for DNA sliding., Mol Cell. 2000 May;5(5):889-95. Print.
- ↑ Voet, Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. p. 578-579. Print.
- ↑ 3.0 3.1 PDBe., European Molecular Biology Laboratory. 2011. Webpage.
- ↑ Newman, Matthew, Structure of BamHI Endonuclease Bound to DNA: Partial Folding and Unfolding on DNA Binding., Science. 1995 Aug;4(269):656-63. Print.
Proteopedia Page Contributors and Editors (what is this?)
Shane Michael Evans, Michal Harel, Alexander Berchansky, Ann Taylor