Inositol 1,4,5-Trisphosphate Receptor
From Proteopedia
| (26 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1n4K' size='350' side='right' caption='Mouse inositol triphosphate receptor ligand-binding core complex with its ligand inositol triphosphate (PDB entry [[1n4k]])' scene='38/382942/Cv/1'> | |
| - | [[Inositol 1,4,5-Trisphosphate Receptor]] binding protein is a ubiquitous protein involved in the Ca<sup>2+</sup> signalling processes in a variety of organisms <ref name="mainpaper">PMID:12442173</ref> | + | ==Introduction== |
| - | + | [[Inositol 1,4,5-Trisphosphate Receptor]] binding protein is a ubiquitous protein involved in the Ca<sup>2+</sup> signalling processes in a variety of organisms <ref name="mainpaper">PMID:12442173</ref>. See also [[Endoplasmic reticulum/Sarcoplasmic reticulum receptors]], [[Receptor]] and [[Ca2+ signalling processes]]. | |
| - | + | ||
| - | + | ||
| - | + | ==Structure== | |
| + | The specific type of inositol 1,4,5-trisphosphate receptor (InsP<sub>3</sub>R) protein discussed here is the mouse type 1 InsP<sub>3</sub>R, also called InsP<sub>3</sub>R1. This polypeptide contains three major regions: the <scene name='38/382942/N_terminal_domain/1'>amino terminal</scene> inositol 1,4,5-trisphosphate (InsP<sub>3</sub>) binding region, the central modulatory region, and the <scene name='38/382942/C_terminal_domain/1'>carboxy-terminus channel region</scene>.<ref name="mainpaper"/> The protein forms an L-shaped structure composed of two asymmetric domains perpendicular to each other.<ref name="mainpaper"/> The N-terminal domain is made up of 12 β-strands and 2 single-turn helices, which come together to form a barrel.<ref name="mainpaper"/> The C-terminal end is quite different, consisting of a bundle made of eight α-helices.<ref name="mainpaper"/> The interface of the two domains is lined with basic residues and forms the <scene name='38/382942/Ip3_binding_pocket/1'>receptor site</scene> for InsP<sub>3</sub>.<ref name="mainpaper"/> The InsP<sub>3</sub>R protein does not belong to a superfamily of proteins. The receptor is thought to span the membrane 6 times, leaving the C-terminus in the cytoplasm.<ref name="functionref"/> | ||
=== Domain Structure === | === Domain Structure === | ||
| - | |||
The protein fold of the β-domain can also be called the β-trefoil. This element is present in other proteins as well, including fibroblast growth factors and mannose receptors.<ref name="mainpaper"/> In the case of the InsP<sub>3</sub>R β-trefoil, the structure was found to be very similar to the β-trefoil of the mannose receptor.<ref name="mainpaper"/> In the β-domain of InsP<sub>3</sub>R1, three of six two-stranded hairpins come together to form a barrel and the other three form a triangular cap for the barrel.<ref name="mainpaper"/> | The protein fold of the β-domain can also be called the β-trefoil. This element is present in other proteins as well, including fibroblast growth factors and mannose receptors.<ref name="mainpaper"/> In the case of the InsP<sub>3</sub>R β-trefoil, the structure was found to be very similar to the β-trefoil of the mannose receptor.<ref name="mainpaper"/> In the β-domain of InsP<sub>3</sub>R1, three of six two-stranded hairpins come together to form a barrel and the other three form a triangular cap for the barrel.<ref name="mainpaper"/> | ||
| Line 18: | Line 16: | ||
=== Binding the InsP<sub>3</sub> Ligand: Mechanism and Structural Components=== | === Binding the InsP<sub>3</sub> Ligand: Mechanism and Structural Components=== | ||
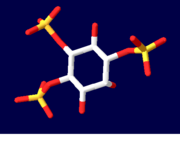
| - | The InsP<sub>3</sub> <scene name='Sandbox_170/1n4k/8'>ligand</scene> sits between the two domains of the protein. Highly basic amino acid residues are present on both domains and are responsible for the binding of InsP<sub>3</sub> to InsP<sub>3</sub>R.<ref name="mainpaper"/> Since the InsP<sub>3</sub> ligand is highly charged, it is very likely to interact with the positively charged amino acids present in the N-terminus InsP<sub>3</sub>-binding domain.<ref name="functionref"/> In binding, water molecules are involved in hydrogen bonding between InsP<sub>3</sub> and its receptor as well as interactions between protein side chains and phosphorous.<ref name="mainpaper"/> | + | The InsP<sub>3</sub> <scene name='Sandbox_170/1n4k/8'>ligand</scene> sits between the two domains of the protein. Highly <scene name='38/382942/Ip3_binding_pocket/1'>basic amino acid residues</scene> are present on both domains and are responsible for the binding of InsP<sub>3</sub> to InsP<sub>3</sub>R.<ref name="mainpaper"/> Since the InsP<sub>3</sub> ligand is highly charged, it is very likely to interact with the positively charged amino acids present in the N-terminus InsP<sub>3</sub>-binding domain.<ref name="functionref"/> In binding, water molecules are involved in hydrogen bonding between InsP<sub>3</sub> and its receptor as well as interactions between protein side chains and phosphorous.<ref name="mainpaper"/> <scene name='38/382942/Cv/4'>Active site</scene> (water molecules shown as red spheres). Coordination of phosphorous groups is mediated by residues in both the β-domain and α-domain. The hydroxyl groups of InsP<sub>3</sub> play a small role in binding to InsP<sub>3</sub>.<ref name="mainpaper"/> Additionally, 9 out of 12 Arg/Lys residues play a very important role in ligand binding and salt bridges to stabilize between the domain regions.<ref name="mainpaper"/> The non-basic residues T266, T267, G268, and Y567 are also integral in InsP<sub>3</sub> coordination: if T267, G268 or Y567 residues are mutated then there will be a significant reduction in ligand binding.<ref name="mainpaper"/> In all likelihood, the InsP<sub>3</sub>-binding site has been found to be made up of multiple sequences present throughout the N-terminal area of the protein.<ref name="functionref"/> This makes the tertiary structure of the protein and proper folding absolutely integral to the function: if the protein does not fold correctly, then the multiple sequences of the protein making up the binding region cannot come together to be at all functional in binding the InsP<sub>3</sub> ligand. |
[[Image:Ligand1.PNG| thumb|Inositol 1,4,5-trisphosphate]] | [[Image:Ligand1.PNG| thumb|Inositol 1,4,5-trisphosphate]] | ||
| Line 37: | Line 35: | ||
The InsP<sub>3</sub>R protein can autophosphorylate itself and is a substrate for multiple protein kinases.<ref name="functionref"/> These kinases include cyclic AMP-dependent protein kinase (PKA), cyclic GMP-dependent protein kinase (PKG) and others.<ref name="functionref"/> The protein kinases are thought to interact with the InsP<sub>3</sub> receptor by controlling the sensitivity to Ca<sup>2+</sup> in different tissues as well as affecting the sensitivity of InsP<sub>3</sub> itself to Ca<sup>2+</sup>.<ref name="functionref"/> | The InsP<sub>3</sub>R protein can autophosphorylate itself and is a substrate for multiple protein kinases.<ref name="functionref"/> These kinases include cyclic AMP-dependent protein kinase (PKA), cyclic GMP-dependent protein kinase (PKG) and others.<ref name="functionref"/> The protein kinases are thought to interact with the InsP<sub>3</sub> receptor by controlling the sensitivity to Ca<sup>2+</sup> in different tissues as well as affecting the sensitivity of InsP<sub>3</sub> itself to Ca<sup>2+</sup>.<ref name="functionref"/> | ||
| + | </StructureSection> | ||
| + | ===3D structures of inositol 1,4,5-trisphosphate receptor=== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[6dqj]], [[6dr2]], [[6dra]], [[6uqk]] – hInsP3R III – human - Cryo EM<br /> | ||
| + | [[6dqn]], [[6dqs]], [[6dqv]], [[6dqz]], [[6dr0]], [[6drc]] – hInsP3R III + IP3 - Cryo EM<br /> | ||
| + | [[5x9z]], [[5xa0]] – mInsP3R I - mouse<br /> | ||
| + | [[3jrr]] – mInsP3R III ligand-binding suppressor domain <br /> | ||
| + | [[1xzz]] - mInsP3R I ligand-binding suppressor domain<br /> | ||
| + | [[1n4k]] - mInsP3R I receptor-binding core + IP3<br /> | ||
| + | [[5gug]], [[5xa1]] – mInsP3R I + IP3<br /> | ||
| + | [[3t8s]], [[3uj4]] - rInsP3R I ligand-binding domain - rat<br /> | ||
| + | [[3uj0]] - rInsP3R I ligand-binding domain + IP3<br /> | ||
| + | [[3jav]], [[6mu2]] – rInsP3R I – Cryo EM<br /> | ||
| + | [[7lhe]], [[7lhf]] – rInsP3R I + lipid – Cryo EM<br /> | ||
| + | [[6mu1]] – rInsP3R I + adenophostin – Cryo EM<br /> | ||
==References== | ==References== | ||
<references /> | <references /> | ||
| + | |||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of inositol 1,4,5-trisphosphate receptor
Updated on 19-January-2022
6dqj, 6dr2, 6dra, 6uqk – hInsP3R III – human - Cryo EM
6dqn, 6dqs, 6dqv, 6dqz, 6dr0, 6drc – hInsP3R III + IP3 - Cryo EM
5x9z, 5xa0 – mInsP3R I - mouse
3jrr – mInsP3R III ligand-binding suppressor domain
1xzz - mInsP3R I ligand-binding suppressor domain
1n4k - mInsP3R I receptor-binding core + IP3
5gug, 5xa1 – mInsP3R I + IP3
3t8s, 3uj4 - rInsP3R I ligand-binding domain - rat
3uj0 - rInsP3R I ligand-binding domain + IP3
3jav, 6mu2 – rInsP3R I – Cryo EM
7lhe, 7lhf – rInsP3R I + lipid – Cryo EM
6mu1 – rInsP3R I + adenophostin – Cryo EM
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 Bosanac I, Alattia JR, Mal TK, Chan J, Talarico S, Tong FK, Tong KI, Yoshikawa F, Furuichi T, Iwai M, Michikawa T, Mikoshiba K, Ikura M. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature. 2002 Dec 12;420(6916):696-700. Epub 2002 Nov 17. PMID:12442173 doi:10.1038/nature01268
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 Patel S, Joseph SK, Thomas AP. Molecular properties of inositol 1,4,5-trisphosphate receptors. Cell Calcium. 1999 Mar;25(3):247-64. PMID:10378086 doi:10.1054/ceca.1999.0021
Proteopedia Page Contributors and Editors (what is this?)
Shannon King, Alexander Berchansky, Michal Harel, Ann Taylor, David Canner, Andrea Gorrell, Jaclyn Gordon