AChE inhibitors and substrates
From Proteopedia
(Difference between revisions)
| (32 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | __NOTOC__ | |
| - | + | <StructureSection load='2ace' size='450' side='right' scene='2ace/Com_view/1' caption='Torpedo californica acetylcholinesterase complex with acetylcholine, [[2ace]]' > | |
| - | + | ||
| - | + | ||
==AChE substrate== | ==AChE substrate== | ||
| - | '''Dear readers, this page presents only a small part of the great world of the acetylcholinesterase inhibitors. So, please see also our pages [[AChE inhibitors and substrates (Part II)]], [[AChE inhibitors and substrates (Part III)]], [[AChE bivalent inhibitors]] and [[AChE bivalent inhibitors (Part II)]].''' | ||
| - | The images at the left and at the right correspond to one representative acetylcholinesterase with substrate structure, ''i.e.'' crystal structure of ''Torpedo californica'' acetylcholinesterase with acetylcholine ([[2ace]]). | ||
| - | Solution of the three-dimensional (3D) structure | + | Solution of the three-dimensional (3D) structure of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] (''Tc''AChE) in 1991 <ref name="Sussman">PMID:1678899</ref> opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation. The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis <ref name="Botti">PMID:10545346</ref>. This led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as |
| - | of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] (''Tc''AChE) | + | |
| - | in 1991 <ref name="Sussman">PMID:1678899</ref> opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation. The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis <ref name="Botti">PMID:10545346</ref>. This led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as | + | |
[http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression | [http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression | ||
| - | systems. [http://en.wikipedia.org/wiki/Acetylcholinesterase Acetylcholinesterase] [http://en.wikipedia.org/wiki/Hydrolysis hydrolysizes] the [http://en.wikipedia.org/wiki/Neurotransmitter neurotransmitter] [http://en.wikipedia.org/wiki/Acetylcholine acetylcholine] <scene name='2ace/Cv/2'>(ACh)</scene>, producing <scene name='2ace/Cv/3'>choline and an acetate</scene> group. ACh directly binds <scene name=' | + | systems. [http://en.wikipedia.org/wiki/Acetylcholinesterase Acetylcholinesterase] [http://en.wikipedia.org/wiki/Hydrolysis hydrolysizes] the [http://en.wikipedia.org/wiki/Neurotransmitter neurotransmitter] [http://en.wikipedia.org/wiki/Acetylcholine acetylcholine] <scene name='2ace/Cv/2'>(ACh)</scene>, producing <scene name='2ace/Cv/3'>choline and an acetate</scene> group. ACh directly binds <scene name='22/22/Cv/1'>Ser200</scene> (via its [http://en.wikipedia.org/wiki/Nucleophile nucleophilic] Oγ atom) within the <scene name='2ace/Cv/5'>catalytic triad (Ser200, His440, and Glu327)</scene> (ACh/''Tc''AChE structure [[2ace]]). The residues <scene name='2ace/Cv/6'>Trp84 and Phe330</scene> are also important in the [http://en.wikipedia.org/wiki/Ligand ligand] recognition <ref name="Raves">PMID:8989325</ref>. After this binding acetylcholinesterase <scene name='2ace/Cv/7'>hydrolysizes</scene> ACh. |
==AChE monovalent inhibitors== | ==AChE monovalent inhibitors== | ||
| Line 19: | Line 13: | ||
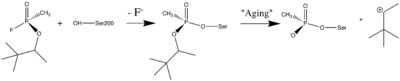
| + | [http://en.wikipedia.org/wiki/Organophosphorus Organophosphorus] (OP) [http://en.wikipedia.org/wiki/Acid_anhydride acid anhydride] [http://en.wikipedia.org/wiki/Nerve_agent nerve agents] are potent inhibitors which rapidly phosphonylate AChE and then may undergo an internal dealkylation reaction (called "aging") to produce an OP-enzyme conjugate that cannot be reactivated. | ||
| + | |||
| + | [[Image:Soman_reaction.png | left | thumb | 400px | Reaction between Ser200Oγ and Soman, assuming an in-line attack by the Oγ, followed by spontaneous dealkylation of the O-pinacolyl group.]] | ||
| + | <br style="clear:both;"/> | ||
| + | |||
| + | |||
| + | ==== Soman ==== | ||
| + | As was mentioned above, AChE hydrolysizes the neurotransmitter <scene name='2wfz/Al/2'>ACh</scene>, producing <scene name='2wfz/Al/3'>choline and an acetate</scene> group. <scene name='2wfz/Al/2'>ACh</scene> directly binds catalytic <scene name='2wfz/Al/4'>Ser200</scene> (via its nucleophilic Oγ atom). <scene name='2wfz/Al/5'>Soman</scene>, [http://en.wikipedia.org/wiki/Soman O-(1,2,2-trimethylpropyl) methylphosphonofluoridate] (<span style="color:violet;background-color:black;font-weight:bold;">fluorine atom is colored violet</span> and <font color='darkmagenta'><b>phosphorus atom is colored darkmagenta</b></font>), is one of the most toxic OPs. Soman inhibits AChE by <scene name='2wfz/Al/6'>covalent binding</scene> to catalytic Ser200, <scene name='2wfz/Al/7'>mimicking ACh</scene>. This process <scene name='2wfz/Al/8'>(phosphonylation)</scene> implicates nucleophilic attack of the Ser200 nucleophilic Oγ atom on the phosphorus atom of soman, with concomitant departure of its fluoride atom. After that AChE catalyzes the <scene name='2wfz/Al/9'>dealkylation ("aging")</scene> of the soman or other OP. This causes irreversible inhibition of AChE, "aged" soman/AChE conjugate can not be reactivated. However, before “aging”, at the step of <scene name='2wfz/Al/8'>phosphonylation</scene>, AChE can be <scene name='2wfz/Al/11'>reactivated</scene> by nucleophiles, such as pralidoxime (2-PAM), resulting in <scene name='2wfz/Al/12'>cleavage</scene> of the phosphonyl adduct from Ser200 Oγ. | ||
| + | At the <scene name='2wfz/Ali/3'>active site of the nonaged soman/TcAChE conjugate</scene> ([[2wfz]]) the catalytic His440 forms hydrogen bonds with Ser200 Oγ and Glu327 Oε1 via its Nε2 and Nδ1 nitrogens, respectively. The O2 atom of soman is within hydrogen bonding distance of His440 Nε2. Soman O1 mimicks carbonyl oxygen of ACh. A water molecule 1001 interacting with soman O2 is represented as a <font color='red'><b>red ball</b></font>. The active site residues of the nonaged soman/TcAChE are colored <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. The O2 atom of the <scene name='2wfz/Ali/4'>dealkylated (aged) soman</scene> ([[2wg0]]) forms a salt bridge with His440 Nε2. The active site residues of the aged soman/TcAChE are colored <span style="color:pink;background-color:black;font-weight:bold;">pink</span>. <scene name='2wfz/Ali/5'>Alignment</scene> of the structures of the nonaged ([[2wfz]]) and aged ([[2wg0]]) conjugates reveals a small, but important, change within the active site - the imidazole ring of His440 is tilted back to a native-like conformation after dealkylation. The water molecule 1001, which interacts with soman O2 in the nonaged crystal structure, is not within hydrogen bonding distance of O2 in the aged crystal structure. 2-PAM binds poorly to the nonaged phosphonylated enzyme (its electron density was not found) and binds in an <scene name='2wfz/Ali/7'>unfavorable and nonfunctional conformation</scene> after soman aging to ''Tc''AChE ([[2wg1]]) <ref name="Sanson">PMID:19642642</ref>. | ||
| + | |||
| + | |||
| + | |||
| + | To understand the basis for [http://en.wikipedia.org/wiki/Enzyme_inhibitor#Irreversible_inhibitors irreversible inhibition], the <scene name='1som/Ache_soman/1'>structure of the aged conjugate</scene> obtained by reaction of ''Tc''AChE with soman was solved by X-ray crystallography to 2.2Å resolution ([[1som]]). The highest positive difference density peak corresponded to the OP phosphorus and was located within covalent bonding distance of the active-site serine (S200). The <scene name='1som/Soman_active_site/3'>OP-oxygen atoms</scene> are within hydrogen-bonding distance of four potential donors from catalytic subsites of the enzyme, suggesting that electrostatic forces significantly stabilize the aged enzyme. The methyl group of soman occupies the <scene name='1som/Soman_acyl_binding/2'>acyl binding pocket</scene>, bounded by Trp233, Phe288, and Phe290 <ref name="Millard">PMID:10353814</ref>. | ||
| + | |||
| + | ====Sarin==== | ||
| + | <scene name='1cfj/Cv/1'>Sarin.</scene> [http://en.wikipedia.org/wiki/Sarin Sarin], O-''iso''propylmethylphosponofluoridate, is an other toxic OP compound. It is also inhibits AChE by covalent binding to the catalytic Ser200. The active sites of aged <scene name='1cfj/Cv/2'>sarin-TcAChE</scene> ([[1cfj]]) and aged soman-TcAChE ([[1som]] and [[2wg0]]) are almost identical and provided structural models for the negatively charged, tetrahedral intermediate that occurs during deacylation with the ACh. | ||
| + | There are four hydrogen bond donors <font color='red'><b>(red dotted lines)</b></font> to the anionic phosphonyl oxygen atoms: the backbone amide nitrogen atoms of Ala201, Gly118, and Gly119, as well as His440 Nε2. The sarin <span style="color:cyan;background-color:black;font-weight:bold;">methyl carbon (colored cyan)</span> is within non-bonded contact distances '''(black dotted lines)''' of Phe288 and Phe290 in the acyl binding pocket <ref name="Millard"/>. | ||
| + | |||
| + | |||
| + | ====DFP==== | ||
| + | <scene name='2dfp/Cv/3'>DFP</scene>, di''iso''propylphosphorofluoridate, is an other toxic OP nerve agent. It is also inhibits AChE by covalent binding to the catalytic Ser200. As in the case with soman ([[1som]]) and sarin ([[1cfj]]), there are four hydrogen bond donors (dotted lines) to the anionic phosphonyl oxygen atoms: the backbone amide nitrogen atoms of Ala201, Gly118, and Gly119, as well as His440 Nε2 at the <scene name='2dfp/Cv/4'>active site</scene> of aged DFP-TcAChE ([[2dfp]]). Phosphorylation with DFP caused an unexpected distortion in the main chain of a loop that includes residues F288 and F290 of the TcAChE acyl binding pocket. F288 and F290 move significantly in the <span style="color:lime;background-color:black;font-weight:bold;">DFP-TcAChE structure (green)</span>, in comparison to their positions in the <font color='magenta'><b>native enzyme</b></font> ([[2ace]]). This is the first major conformational change reported in the active site of any AChE−ligand complex, and it offers a structural explanation for the substrate selectivity of AChE <ref name="Millard"/>. | ||
{{Clear}} | {{Clear}} | ||
| - | [http://en.wikipedia.org/wiki/ | + | ===Treatment of Alzheimer's disease=== |
| + | {{Clear}} | ||
| + | [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease] (AD) is a disorder that attacks the [http://en.wikipedia.org/wiki/Central_nervous_system central nervous system] through progressive degeneration of its neurons. Patients with this disease develop [http://en.wikipedia.org/wiki/Dementia dementia] which becomes more severe as the disease progresses. It was suggested that symptoms of AD are caused by decrease of activity of [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Neocortex neocortical] and [http://en.wikipedia.org/wiki/Hippocampus hippocampal] neurons. Treatment of AD by ACh precursors and [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Agonist agonists] was ineffective or caused severe side effects. ACh hydrolysis by AChE causes termination of cholinergic neurotransmission. Therefore, compounds which inhibit AChE might significantly increase the levels of ACh depleted in AD. Indeed, it was shown that [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor AChE inhibitors] improve the cognitive abilities of AD patients at early stages of the disease development. The first generation of AD drugs were AChE inhibitors: alcaloids like [http://en.wikipedia.org/wiki/Huperzine_A (-)-Huperzine A (HupA)] and [http://en.wikipedia.org/wiki/Galantamine (-)-galanthamine (GAL, Reminyl)]; [http://en.wikipedia.org/wiki/Chemical_synthesis synthetic] compounds [http://en.wikipedia.org/wiki/Tacrine tacrine (Cognex)] and [http://en.wikipedia.org/wiki/Rivastigmine rivastigmine (Exelon)]. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====Donepezil (Aricept)==== | ||
| + | [[Donepezil]] is a potent [[Acetylcholinesterase]] (AChE) inhibitor to the active site of <scene name='29/2908/1eve_e20_cartoon/3'>AChE</scene>. By inhibiting AChE, the important neurotransmitter, [[acetylcholine]], is degraded at a slower rate, helping reverse the marked decrease in neuronal function evident in [[Alzheimer's Disease]] patients. Donepezil binds along the active-site gorge, extending from the anionic subsite <scene name='Donepezil/Trp_84/1'>near Trp 84</scene> to the peripheral anionic site <scene name='Donepezil/Trp_279/1'>near Trp 279</scene>. Interestingly, it does not directly interact with the catalytic triad of acetylcholinesterase nor the oxyanion hole. Further, donepezil does not form any direct hydrogen bonds with AChE nor electrostatic interactions, but rather only interacts via aromatic stacking and solvent mediated interactions. It <scene name='Donepezil/Bound/1'>primarily interacts</scene> with Glu 199, His 440, Phe 330, Trp 84, Tyr 334, Tyr 121, Phe 331, Phe 288, Ser 286, Phe 290, Arg 289, Trp 279, & Leu 282 to tightly bind to AChE.<ref name="Kryger">PMID:10368299</ref>. For more details see [[Aricept_Complexed_with_Acetylcholinesterase]]. | ||
| - | [[Image:Soman_reaction.png | left | thumb | 800px | Reaction between Ser200Oγ and Soman, assuming an in-line attack by the Oγ, followed by spontaneous dealkylation of the O-pinacolyl group.]] | ||
| - | <br style="clear:both;"/> | ||
{{Clear}} | {{Clear}} | ||
| - | + | ====(-)-Huperzine A==== | |
| + | [http://en.wikipedia.org/wiki/Huperzine_A (-)-Huperzine A], discovered by Chinese scientists from 1980s, has been proved to be a powerful, highly specific, and [http://en.wikipedia.org/wiki/Enzyme_inhibitor#Reversible_inhibitors reversible inhibitor] of AChE. It is a novel [http://en.wikipedia.org/wiki/Alkaloid alkaloid] originally isolated from the '''Traditional Chinese medicine''' [http://en.wikipedia.org/wiki/Traditional_Chinese_medicine] Qian Ceng Ta which is produced from the whole plant of the [http://en.wikipedia.org/wiki/Firmoss firmoss][http://en.wikipedia.org/wiki/Huperzia_serrata ''Huperzia serrata'']. Qian Ceng Ta has been used for over 1000 years in China for treatment of [http://en.wikipedia.org/wiki/Bruise contusions], [http://en.wikipedia.org/wiki/Strain_(injury) strains], [http://en.wikipedia.org/wiki/Swelling_(medical) swellings], [http://en.wikipedia.org/wiki/Schizophrenia schizophrenia] and [http://en.wikipedia.org/wiki/Myasthenia_gravis myasthenia gravis]. Shuangyiping[http://www.54md.com/drugstore/pic/gpic_25fd25197010a0fb4a680516735e613c.jpg], a tablet form of HupA produced from the extracts of ''Huperzia serrata'', was developed in 1996 as a new drug for symptomatic treatment of Alzheimer’s disease in China. Compared with the other three [http://en.wikipedia.org/wiki/Food_and_Drug_Administration_(United_States) FDA]-approved drugs for the treatment of Alzheimer’s disease, Donepezil (Aricept), Rivastigmine (Exelon), Galanthamine (Reminyl), HupA has better penetration through the [http://en.wikipedia.org/wiki/Blood-brain_barrier blood-brain barrier], higher oral [http://en.wikipedia.org/wiki/Bioavailability bioavailability], and longer duration of AChE inhibitory action. The structure of HupA shows some similarity to other known AChE inhibitors. The molecule is fairly rigid and contains an [http://en.wikipedia.org/wiki/Aromaticity aromatic] system as well as a [http://en.wikipedia.org/wiki/Amine primary amino group] that is probably [http://en.wikipedia.org/wiki/Protonation protonated] at physiological [http://en.wikipedia.org/wiki/PH pH]. Various suggestions have been made with respect to its orientation within the active site of AChE, and with respect to the amino acid residue with which its putative [http://en.wikipedia.org/wiki/Pharmacophore pharmacophoric] groups might interact. Solution of the 3D structure of a complex of HupA with AChE would permit unequivocal resolution of this issue and it would also provide a rational basis for structure-related [http://en.wikipedia.org/wiki/Drug_design drug design] aimed at developing synthetic analogues of HupA with improved therapeutic properties. | ||
| + | The [http://en.wikipedia.org/wiki/X-ray_crystallography crystal structure] of the complex of ''Tc''AChE with HupA at 2.5 Å resolution ([[1vot]]) was determined in 1997 and it shows an unexpected orientation for the inhibitor with surprisingly few strong direct interactions with protein residues to explain its high affinity. <font color='blueviolet'><b>HupA</b></font> binds to ''Tc''AChE at the active site, and its <scene name='1vot/Active_site/8'>observed orientation is almost orthogonal</scene> in comparison to <font color='gray'><b>ACh</b></font>. The principal interactions of <scene name='1vot/1vot_ache_interactions/2'>HupA with TcAChE</scene> are including: a direct <scene name='1vot/1vot_199_130_117/2'>hydrogen bond with Tyr130 and HBs with Glu199 and Gly117</scene> <span style="color:orange;background-color:black;font-weight:bold;">(colored orange)</span> through a water molecule as a linker at the bottom of the gorge; [http://en.wikipedia.org/wiki/Cation-pi_interaction cation-π] interactions between the amino group of <scene name='1vot/1vot_84_330/2'>HupA and Trp84 and Phe330</scene> <span style="color:lime;background-color:black;font-weight:bold;">(colored green)</span> with the distance between the nitrogen and the centroid of the aromatic rings of 4.8 and 4.7 Å, respectively; at the top of the gorge, [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonds] through two water molecules as linkers formed between the amino group of <scene name='1vot/1vot_70_72_81_85_121/3'>HupA and Tyr70, Asp72, Ser81, Asn85 and Tyr121</scene> <font color='magenta'><b>(colored magenta)</b></font>. An unusually short (~3.0 Å) C-H→O HB has been seen between the ethylidene methyl group of <scene name='1vot/1vot_440/2'>HupA and the main chain oxygen of His440</scene> <font color='crimson'><b>(colored crimson)</b></font> <ref name="Raves">PMID:8989325</ref>. | ||
| - | + | {{Clear}} | |
| - | + | ||
| - | + | ||
| + | ====Tacrine==== | ||
| + | <scene name='AChE_inhibitors_and_substrates/Com_view_tacrine/1'>Tacrine</scene> [http://en.wikipedia.org/wiki/Tacrine]. In the X-ray crystal structure of ''Tc''AChE/<scene name='AChE_inhibitors_and_substrates/Com_view_tacrine/2'>tacrine</scene> complex which was determined at 2.8 Å resolution, the tacrine is seen <font color='magenta'><b>(magenta)</b></font> bound in the active site of ''Tc''AChE ([[1acj]]) <ref name="Harel">PMID:8415649</ref>. <font color='gray'><b>ACh (gray)</b></font> is shown for comparison. In the crystal structure of ''[http://en.wikipedia.org/wiki/Torpedo_californica Torpedo californica]'' [[acetylcholinesterase]] (''Tc''AChE) complexed with [http://en.wikipedia.org/wiki/Tacrine tacrine] (THA), THA's [http://en.wikipedia.org/wiki/Acridine acridine] ring is stacked between the [http://en.wikipedia.org/wiki/Aromaticity aromatic rings] of <scene name='80/80578/Cv/1'>W84 and F330</scene>, near the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] of <scene name='1acj/Active_site_of_1acj/2'>AChE's active site</scene> which consists of '''S200''', '''E327''', '''H440'''. When comparing 3 recent complexes of ''Tc''AChE, i.e. edrophonium (EDR), decamethonium (DECA) and THA, the only major conformational difference between them is seen in the orientation of the [http://en.wikipedia.org/wiki/Phenyl_group phenyl ring] of F330. In the DECA complex it lies parallel to the surface of the gorge; in the other two complexes it is positioned to make contact with the bound ligand. This close interaction was confirmed by photoaffinity labeling by a 3H-labeled photosensitive probe, which labeled, predominantly, F330 within the active site. Labeling of <scene name='1acj/Tha_active_site_test_2/2'>W279</scene> was also observed. One mole of label is incorporated per mole of AChE inactivated, indicating that labeling of W279 and that of F330 are mutually exclusive. The structural and chemical data, together, show the important role of aromatic groups as binding sites for quaternary ligands, and they provide complementary evidence assigning W84 and F330 to the "anionic" subsite of the active site and W279 to the "peripheral" anionic site. | ||
{{Clear}} | {{Clear}} | ||
| - | + | ====Huprine X==== | |
| + | <scene name='1e66/Active_site/1'>Huprine X</scene> ('''HUP'''erzine A + tac'''RINE''') is one of the most potent [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor#Reversible_inhibitor reversible AChE inhibitors]. This <scene name='1e66/Active_site/2'>synthetic hybrid</scene> consists of a carbobicyclic moiety resembling that of (−)-<scene name='1e66/Active_site/12'>huperzine A</scene> <font color='blueviolet'><b>(colored blueviolet)</b></font> and the [http://en.wikipedia.org/wiki/4-Aminoquinoline 4-aminoquinoline] substructure of <scene name='1e66/Active_site/7'>tacrine</scene> <font color='magenta'><b>(colored magenta)</b></font>. Both these compounds are known AChE inhibitors. <font color='blueviolet'><b>(−)-Huperzine A</b></font> and <font color='magenta'><b>tacrine</b></font> positions partially overlap each other at the ''Tc''AChE <scene name='1e66/Active_site/13'>active site</scene>. ''Tc''AChE residues interacting with (−)-huperzine A ([[1vot]]) <span style="color:orange;background-color:black;font-weight:bold;">are colored orange</span> and with tacrine ([[1acj]]) <span style="color:cyan;background-color:black;font-weight:bold;">are colored cyan</span>. The <scene name='1e66/Active_site/10'>conformation</scene> of the 4-aminoquinoline substructure of the huprine X in its complex with ''Tc''AChE ([[1e66]], ''Tc''AChE interacting residues are in <span style="color:lime;background-color:black;font-weight:bold;">green</span>) is very similar to that of tacrine. The ring system of (−)-huperzine A is <scene name='1e66/Active_site/14'>rotated</scene> almost 180° relative to that of huprine X <ref name="Dvir">PMID:11863435</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====Galanthamine==== | ||
| + | <scene name='AChE_inhibitors_and_substrates/Com_view_gal/1'>Galanthamine (GAL)</scene> [http://en.wikipedia.org/wiki/Galantamine]. <scene name='AChE_inhibitors_and_substrates/Com_view_gal/2'>GAL</scene> <font color='red'><b>(red)</b></font> is an [http://en.wikipedia.org/wiki/Alkaloid alkaloid] from the flower snowdrop ([http://en.wikipedia.org/wiki/Galanthus ''Galanthus nivalis'']). The [http://en.wikipedia.org/wiki/X-ray_crystallography X-ray crystal structure] of the ''Tc''AChE/GAL complex ([[1dx6]]) was determined at 2.3 Å resolution. The inhibitor binds at the base of the [http://en.wikipedia.org/wiki/Active_site active site] gorge of ''Tc''AChE, interacting with both the choline-binding site (Trp84) and the acyl-binding pocket (Phe288, Phe290). The [http://en.wikipedia.org/wiki/Amine tertiary amine] appears to make a non-conventional [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond], via its N-methyl group, to Asp72. The [http://en.wikipedia.org/wiki/Hydroxyl#Hydroxyl_group hydroxyl group] of the inhibitor makes a strong hydrogen bond (2.7 Å) with Glu199 <ref name="Greenblatt">PMID:10606746</ref>. <font color='gray'><b>ACh (gray)</b></font> is shown for comparison. | ||
{{Clear}} | {{Clear}} | ||
| - | ==== | + | ====Edrophonium==== |
| - | <scene name=' | + | <scene name='2ack/Com_view/1'>Edrophonium (EDR)</scene> [http://en.wikipedia.org/wiki/Edrophonium] is stacked between the [http://en.wikipedia.org/wiki/Aromatic aromatic rings] of <scene name='2ack/Com_view/2'>W84 and F330</scene>, near the ''Tc''AChE <scene name='2ack/Com_view/3'>catalytic triad</scene> which consists of <font color='magenta'><b>'''S200'''</b></font>, <font color='magenta'><b>'''E327'''</b></font>, and <font color='magenta'><b>'''H440'''</b></font> ([[2ack]] or [[1ax9]]) <ref name="Ravelli">PMID:10089512</ref>. |
| - | + | ||
| + | ====Galanthamine iminium derivative==== | ||
| + | The X-ray structure of ''Tc''AChE in complex with galanthamine [http://en.wikipedia.org/wiki/Iminium iminium] derivative ('''compound 5''') was determined at 2.05 Å resolution ([[1w6r]]). The <scene name='1w6r/Alignment/6'>binding mode</scene> of this compound <font color='cyan'><b>(cyan)</b></font> with ''Tc''AChE is virtually identical to that of <font color='red'><b>galanthamine (red)</b></font> itself ([[1dx6]]). The ''Tc''AChE residues which interact with galanthamine in the galanthamine/''Tc''AChE complex are colored <font color='pink'><b>pink</b></font>, while those of '''compound 5'''/''Tc''AChE are in <font color='lime'><b>lime</b></font>. The main structural change is the side-chain movement of <font color='lime'><b>Phe330</b></font> in '''compound 5'''/''Tc''AChE complex, in comparison to that of galanthamine. <font color='cyan'><b>Compound 5</b></font> differs from galanthamine by the presence of <scene name='1w6r/Alignment/7'>quaternary nitrogen atom</scene> <font color='blue'><b>(N<sup>+</sup>; blue)</b></font> instead N of <font color='red'><b>galanthamine</b></font>. This chemical difference causes the structural change in ''Tc''AChE and the slight decrease in affinity of '''compound 5''' to ''Tc''AChE in comparison to galanthamine <ref name="Greenblatt">PMID:15563167</ref>. | ||
{{Clear}} | {{Clear}} | ||
| - | ====DFP==== | ||
| - | <scene name='2dfp/Cv/3'>DFP</scene>, di''iso''propylphosphorofluoridate, is an other toxic OP nerve agent. It is also inhibits AChE by covalent binding to the catalytic Ser200. As in the case with soman ([[1som]]) and sarin ([[1cfj]]), there are four hydrogen bond donors (dotted lines) to the anionic phosphonyl oxygen atoms: the backbone amide nitrogen atoms of Ala201, Gly118, and Gly119, as well as His440 Nε2 at the <scene name='2dfp/Cv/4'>active site</scene> of aged DFP-TcAChE ([[2dfp]]). Phosphorylation with DFP caused an unexpected distortion in the main chain of a loop that includes residues F288 and F290 of the TcAChE acyl binding pocket. F288 and F290 move significantly in the <font color='lime'><b>DFP-TcAChE structure (lime)</b></font>, in comparison to their positions in the <font color='magenta'><b>native enzyme</b></font> ([[2ace]]). This is the first major conformational change reported in the active site of any AChE−ligand complex, and it offers a structural explanation for the substrate selectivity of AChE <ref name="Millard"/>. | ||
| - | </ | + | ====Rivastigmine==== |
| + | Rivastigmine (Exelon) is a [http://en.wikipedia.org/wiki/Carbamate carbamate] inhibitor of AChE, and it is currenly used in therapy of [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease]. [http://en.wikipedia.org/wiki/Rivastigmine Rivastigmine (Exelon)] (colored yellow) interacts with ''Tc''AChE <font color='lime'><b>(colored lime)</b></font> at the <scene name='1gqr/Active_site/4'>active-site gorge</scene> ([[1gqr]]). The carbamyl moiety of rivastigmine is <scene name='1gqr/Active_site/9'>covalently bound</scene> to the active-site S200 Oγ. The second part of rivastigmine (the leaving group), NAP ((−)-S-3-[1-(dimethylamino)ethyl]phenol) is also held in the active-site gorge, but it is <scene name='1gqr/Active_site/6'>separated</scene> from the carbamyl moiety, hence, carbamylation took place. The <scene name='1gqr/Active_site/7'>crystal structure</scene> of ''Tc''AChE/<font color='magenta'><b>NAP (colored magenta)</b></font> is known ([[1gqs]]). The <font color='violet'><b>''Tc''AChE active-site residues</b></font> which are interacting with NAP are <font color='violet'><b>colored violet</b></font>. NAP is located in a similar region of ''Tc''AChE active site, but with different orientation than that of the NAP part (colored yellow) in the ''Tc''AChE/rivastigmine complex. Only H440 and F330 significantly change their side-chain conformations. <scene name='1gqr/Active_site/8'>Overlap</scene> of the ''Tc''AChE active sites in 4 different structures (<font color='lime'><b>''Tc''AChE</b></font>/rivastigmine ([[1gqr]]), <font color='violet'><b>''Tc''AChE</b></font>/<font color='magenta'><b>NAP</b></font> ([[1gqs]]), <font color='cyan'><b>native ''Tc''AChE</b></font> ([[2ace]]), and ''Tc''AChE/'''VX''' ([[1vxr]], ''Tc''AChE colored white and VX black) reveals that the conformation of H440 in the ''Tc''AChE/NAP structure is very similar its conformation in the native ''Tc''AChE ([[2ace]]), but the distance between H440 Nδ and E327 Oε is significantly longer in the ''Tc''AChE/rivastigmine and the ''Tc''AChE/'''VX''' complexes. This structural change disrupts the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] consisting of S200, E327, H440. This could explain the very slow kinetics of AChE reactivation after its inhibition by rivastigmine <ref name="Bar-On">PMID:11888271</ref>. | ||
{{Clear}} | {{Clear}} | ||
| + | ====Thioflavin T==== | ||
| + | The ''Tc''AChE active site consists of two binding subsites. One of them is the "catalytic anionic site" (CAS), which involves the catalytic triad <scene name='2j3q/Active_site/2'>Ser200, His440, and Glu327</scene> <font color='orange'><b>(colored orange)</b></font> and the conserved residues <scene name='2j3q/Active_site/3'>Trp84 and Phe330</scene> which also participate in ligand recognition. Another conserved residue <scene name='2j3q/Active_site/4'>Trp279</scene> <font color='cyan'><b>(colored cyan)</b></font> is situated at the second binding subsite, termed the "peripheral anionic site" (PAS), ~14 Å from CAS. <scene name='2j3q/Active_site/6'>Thioflavin T</scene> is a good example of the PAS-binding AChE inhibitors. <scene name='2j3q/Active_site/7'>Superposition</scene> of the crystal structure of the <font color='red'><b>edrophonium</b></font>/''Tc''AChE (mentioned above as a CAS-binding inhibitor) ([[2ack]]) on the <font color='magenta'><b>thioflavin T</b></font>/''Tc''AChE complex structure ([[2j3q]]) shows that these ligands' positions do not overlap. Of note is that Phe330, which is part of the CAS, is the single residue interacting with <font color='magenta'><b>thioflavin T</b></font>. This residue is the only one which significantly <scene name='2j3q/Active_site/9'>changes its conformation</scene> to avoid clashes in comparison to other CAS residues of the <font color='red'><b>edrophonium</b></font>/''Tc''AChE complex <ref name="Ravelli">PMID:10089512</ref> <ref name="Sonoda">PMID:18512913</ref>. | ||
| + | |||
| + | {{Clear}} | ||
| + | ====Methylene blue==== | ||
| + | The photosensitizer, <scene name='Journal:Protein_Science:1/Cv/3'>methylene blue (MB)</scene> <font color='darkmagenta'><b>(colored in darkmagenta)</b></font>, generates singlet oxygen that irreversibly inhibits Torpedo californica acetylcholinesterase (''Tc''AChE). In the dark, it inhibits reversibly. | ||
| + | MB is a noncompetitive inhibitor of ''Tc''AChE, competing with reversible inhibitors directed at both ‘‘anionic’’ subsites, but a single site is involved in inhibition. The crystal structure reveals a <scene name='Journal:Protein_Science:1/Cv1/2'>single MB stacked against Trp279 in the PAS</scene>, oriented down the gorge toward the CAS ([[2w9i]]); it is plausible that irreversible inhibition is associated with photooxidation of this residue and others within the active-site gorge. Superposition of the '''PAS regions''' of the <font color='darkmagenta'><b>MB</b></font>/''Tc''AChE ([[2w9i]]) and <font color='magenta'><b>thioflavin T</b></font>/''Tc''AChE ([[2j3q]]) complexes reveals <scene name='Journal:Protein_Science:1/Cv1/4'>similarity between positions of these ligands</scene>. As the conformation of ''Tc''AChE in the crystal structures of the two complexes is practically identical, only that of the <font color='darkmagenta'><b>MB</b></font>/''Tc''AChE structure ([[2w9i]]) is shown. The kinetic and spectroscopic data showing that inhibitors binding at the '''CAS''' can impede binding of MB are reconciled by docking studies showing that the <scene name='Journal:Protein_Science:1/Cv2/5'>conformation adopted by Phe330</scene>, midway down the gorge, in the MB/''Tc''AChE crystal structure, precludes simultaneous binding of a second MB at the CAS (<font color='blueviolet'><b>2nd MB is colored blueviolet</b></font>, <span style="color:orange;background-color:black;font-weight:bold;">Phe330 of the crystal structure is in orange</span> and <font color='indigo'><b>Phe330 of the modeled structure is in indigo</b></font>). Conversely, binding of ligands at the '''CAS''' dislodges MB from its preferred locus at the '''PAS'''. The data presented demonstrate that TcAChE is a valuable model for understanding the molecular basis of local photooxidative damage.<ref name="Paz">PMID:22674800</ref><ref>PMID:26990888</ref>, and see also: | ||
| + | *[[Journal:Protein_Science:1]] | ||
| + | *[[Journal:Protein_Science:2]] | ||
| + | {{Clear}} | ||
| + | |||
| + | ==AChE bivalent inhibitors== | ||
| + | ====OTMA==== | ||
| + | OTMA is a nonhydrolyzable [http://en.wikipedia.org/wiki/Substrate_analog substrate analogue] of AChE. Its [http://en.wikipedia.org/wiki/Hydrolysis hydrolysis] is impossible as <scene name='2vja/Common/3'>OTMA</scene> possesses <scene name='2vja/Common/4'>carbon</scene> atom instead of the <scene name='2vja/Common/5'>ester oxygen</scene> in the AChE natural substrate ACh. Similarly to ACh, OTMA covalently binds to the ''Tc''AChE ([[2vja]]) <scene name='2vja/Active_site/1'>Ser200</scene> Oγ at the CAS. At this subsite OTMA also interacts with <scene name='2vja/Active_site/2'>Trp84, Phe330</scene> ([http://en.wikipedia.org/wiki/Cation-pi_interaction cation-π interactions]); <scene name='2vja/Active_site/3'>Glu199</scene> (electrostatic interaction); <scene name='2vja/Active_site/4'>Gly118, Gly119, and Ala201</scene> (hydrogen bonds). OTMA binds not only at CAS, but also at PAS. A second OTMA molecule interacts with <scene name='2vja/Active_site/5'>Trp279, Tyr70</scene> (cation-π interactions), and <scene name='2vja/Active_site/6'>Tyr121</scene> (weak hydrogen bond) <ref name="Colletier">PMID:18701720</ref>. Thus, this dual binding mode of OTMA with ''Tc''AChE (to CAS and PAS) could be prototypical for [[AChE bivalent inhibitors]]. | ||
| - | === | + | ===Tacrine- and hupyridone-containing compounds=== |
| + | The <scene name='1zgb/Act_site/3'>active site</scene> of ''Tc''AChE consists of two binding subsites. First of them is the "catalytic anionic site" (CAS), which involves mentioned above [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] <scene name='1zgb/Act_site/8'>Ser200, His440, and Glu327</scene> <font color='orange'><b>(colored orange)</b></font> and the [http://en.wikipedia.org/wiki/Conserved_sequence#Conserved_protein_sequences_and_Structures conserved residues] <scene name='1zgb/Act_site/5'>Trp84</scene> and <scene name='1zgb/Act_site/10'>Phe330</scene> also participating in ligands recognition. Another conserved residue <scene name='1zgb/Act_site/11'>Trp279</scene> <font color='cyan'><b>(colored cyan)</b></font> is situated at the second binding subsite, termed the "peripheral anionic site" (PAS), ~14 Å from CAS. Therefore, the ligands that will be able to interact with both these subsites, will be more potent [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor AChE inhibitors] in comparison to compounds interacting only with CAS. One of the ways to produce such ligands is to introduce two active substances into one compound. If it is spatially necessary these subunits could be divided by alkyl linker with suitable length. | ||
| - | ''' | + | ====(''RS'')-(±)-tacrine-(10)-hupyridone==== |
| - | = | + | According to the strategy of the use of a bivalent ligand, the <scene name='1zgb/Comp/7'>inhibitor</scene> '''(''RS'')-(±)-tacrine-(10)-hupyridone''' ((R)-3 or (S)-3) was designed and synthesized. It consists of mentioned in the page '[[AChE inhibitors and substrates]]' <scene name='1zgb/Comp/8'>tacrine</scene> <font color='magenta'><b>(colored magenta)</b></font>, 10-carbon <scene name='1zgb/Comp/9'>linker</scene> <font color='yellow'><b>(yellow)</b></font>, and <scene name='1zgb/Comp/10'>hupyridone</scene> <font color='red'><b>(red)</b></font>. The tacrine moiety of this inhibitor binds at the CAS, the linker spans the <scene name='1zgb/Act_site/12'>active-site</scene> gorge, and the hupyridone moiety binds at the PAS. |
| + | The comparison of the (R)-3/''Tc''AChE and tacrine/''Tc''AChE complexes at the <scene name='1zgb/Align/2'>active site</scene>. A <scene name='1zgb/Align/7'>comparison</scene> of the trigonal (R)-3/''Tc''AChE structure ([[1zgb]]; <font color='cyan'><b>(R)-3 colored cyan</b></font>; ''Tc''AChE residues interacting with (R)-3 are colored sea-green) with the crystal structure of tacrine/''Tc''AChE ([[1acj]], <font color='magenta'><b>tacrine colored magenta</b></font>; residues interacting with [http://en.wikipedia.org/wiki/Tacrine tacrine] are colored <font color='pink'><b>pink</b></font>) reveals a similar binding mode for the tacrine moiety. In both structures the tacrine ring is situated at the CAS, between the [http://en.wikipedia.org/wiki/Aromaticity aromatic] residues Trp84 and Phe330. Steric clash with the 10-carbon linker could explain the tilt observed for the Phe330 <font color='yellow'><b> (yellow</b></font> and transparent in the tacrine/''Tc''AChE). <font color='red'><b>Water molecules are shown as red spheres.</b></font> | ||
| + | The tacrine unit of (R)-3 N forms <scene name='1zgb/Align/8'>hydrogen bond</scene> with His440 O (3.0 Å) similar to that of tacrine alone. Similarly to the tacrine/''Tc''AChE structure the <font color='red'><b>system of three water molecules</b></font> at the CAS ((R)-3/''Tc''AChE) binds the tacrine-linker N via hydrogen bonds to Ser81 O, Ser122 Oγ, and Asn85 Oδ1 (2.6-3.5 Å). | ||
| + | The <scene name='1zgc/Al/5'>overlap</scene> of <font color='cyan'><b>(R)-3 (cyan)</b></font> and <font color='orange'><b>(S)-3</b></font> ([[1zgc]]) bound to the ''Tc''AChE active site in the orthorhombic forms is shown. <scene name='1zgc/Zgc_h22/2'>Superposition</scene> of <font color='magenta'><b>(S,S)-(-)-4a (magenta)</b></font> and <font color='orange'><b>(S)-3 (orange, orthorhombic ''Tc''AChE)</b></font> demonstrates the similar mode of binding of the hupyridone unit at the PAS. The residues Trp279 (top) and Trp84 (bottom) represent the PAS and the CAS, respectively <ref name="Haviv">PMID:16076210</ref>. | ||
| - | + | {{Clear}} | |
| - | == | + | ====Bis-hupyridone==== |
| - | + | The comparison of the (R)-3/''Tc''AChE ([[1zgb]]) and bis-hupyridone/''Tc''AChE complexes ([[1h22]] and [[1h23]]) at the <scene name='1zgb/Align2/2'>active site</scene>. <scene name='1zgb/Align2/8'>Superposition</scene> of the <font color='cyan'><b>(''R'')-tacrine-(10)-hupyridone ((R)-3, cyan)</b></font> and <font color='orange'><b>(S,S)-(-)-''Bis''(12)-hupyridone ('''(S,S)-(-)-4b''', orange, ''i.e.'' 12-carbon-tether-linked hupyridone dimer)</b></font> and <font color='plum'><b>(S,S)-(-)-''Bis''(10)-hupyridone ('''(S,S)-(-)-4a''', plum)</b></font> complexes demonstrates the binding mode of the hupyridone moiety. <font color='magenta'><b>TcAChE residues of symmetry-related molecule are shown in magenta.</b></font> X-ray structures of ''Tc''AChE complexed with these 10- and 12-carbon-tether-linked 2 <scene name='1zgb/Align2/9'>dimers</scene> <font color='plum'><b>(S,S)-(-)-4a</b></font> and <font color='orange'><b>(S,S)-(-)-4b</b></font> show one subunit bound at the <scene name='1zgb/Align2/10'>CAS</scene>, the linker spanning the gorge, and the other subunit bound at the <scene name='1zgb/Align2/11'>PAS</scene>. | |
| - | + | There are two <scene name='1zgb/Align2/12'>hydrogen bonds</scene> connecting the <font color='cyan'><b>hupyridone</b></font> <font color='red'><b>O</b></font> to <font color='magenta'><b>Lys11</b></font> <font color='blue'><b>Nζ</b></font> and <font color='cyan'><b>hupyridone</b></font> <font color='blue'><b>N</b></font> to <font color='magenta'><b>Gln185</b></font> <font color='red'><b>Oε1</b></font> of a <font color='magenta'><b>symmetry-related molecule</b></font> at <font color='cyan'><b>(R)-3</b></font>/''Tc''AChE complex. <font color='red'><b>Water molecules are shown as red spheres.</b></font> Another hydrogen bond connects the <font color='cyan'><b>hupyridone</b></font> <font color='red'><b>O</b></font> to a water molecule, which is bound to Ser286 N. Similarly, the hupyridone-PAS unit of both <font color='plum'><b>(-)-4a</b></font> and <font color='orange'><b>(-)-4b</b></font> forms direct and an indirect hydrogen bonds with the protein backbone in the PAS region <ref name="Haviv"/> <ref name="Wong">PMID:12517147</ref>. | |
| - | + | {{Clear}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | |||
| + | ====Bis(''n'')-tacrine derivatives==== | ||
| + | <font color='magenta'><b>2d</b></font> and <font color='orange'><b>2f</b></font> are bis(''n'')-tacrine derivatives with n=5 and 7 (number of carbons in the linkers), respectively. These compounds are more potent and selective AChE inhibitors than tacrine alone. The binding of the tacrine moiety of <scene name='2cmf/Comparison/2'>2d</scene> at the ''Tc''AChE catalytic anionic site (CAS) is similar to that of <font color='red'><b>tacrine </b></font> in the tacrine/''Tc''AChE complex ([[1acj]]). The second tacrine moiety of the <font color='magenta'><b>2d</b></font> interacts with the peripheral anionic site (PAS) near Trp279. The interaction of <font color='magenta'><b>2d</b></font> at the CAS causes an increase of the <scene name='2cmf/Comparison/3'>distance</scene> between Ser200 Oγ and H440 Nε2 atoms, and, therefore, disruption of the catalytic triad (Ser200, H220, E327) as seen in the <font color='cyan'><b>native structure</b></font> ([[2ace]]). The binding of 2d results in <scene name='2cmf/Comparison/4'>major structural changes</scene> in the Val281-Ser291 loop changing the surface of the active-site gorge from its <font color='cyan'><b>native conformation</b></font> ([[2ace]]). The tacrine moiety of the compound <font color='orange'><b>'''2f''' (heptylene-linked bis-tacrine</b></font> at the CAS, [[2ckm]]) adopts similar <scene name='2cmf/Comparison/5'>conformation</scene> as tacrine in the tacrine/''Tc''AChE complex and the tacrine moiety of the <font color='magenta'><b>2d</b></font> at the CAS. The second tacrine moiety of the <font color='orange'><b>2f</b></font> interacts with PAS near the Trp279, like <font color='magenta'><b>2d</b></font>. The <scene name='2cmf/Comparison/6'>binding</scene> of <font color='orange'><b>2f</b></font> does not cause significant structural changes in <font color='plum'><b>''Tc''AChE</b></font> from its <font color='cyan'><b>native structure</b></font>. <scene name='2cmf/Overlap/7'>Comparison</scene> of the structures of <font color='magenta'><b>2d</b></font>/''Tc''AChE and <font color='orange'><b>2f</b></font>/''Tc''AChE reveals different contacts between the tacrine moieties of these compounds at the PAS and ''Tc''AChE. There are two additional structures of tacrine-containing ''Tc''AChE complexes: compounds <scene name='2cmf/Comparison/8'>6</scene> ([[1ut6]]) and <scene name='2cmf/Comparison/9'>7</scene> ([[1odc]]). The tacrine moieties of these compounds adopt similar conformations and interactions with CAS as the tacrine in the tacrine/''Tc''AChE, <font color='orange'><b>2f</b></font>/''Tc''AChE and <font color='magenta'><b>2d</b></font>/''Tc''AChE. Inhibitors '''6''' and '''7''' are spanning the <scene name='2cmf/Comparison/10'>active-site gorge</scene> between the CAS and the PAS, but since compound '''7''' lacks the second tacrine moiety, Trp279 adopts a different conformation in this complex structure. In the three structures: native ''Tc''AChE (cyan), '''cmp 6'''/''Tc''AChE complex (white), and '''cmp 7'''/''Tc''AChE complex (crimson) , all the ''Tc''AChE active-site gorge residues have identical conformation except Trp279 <ref name="Harel">PMID:8415649</ref> <ref name="Raves">PMID:8989325</ref> <ref name="Rydberg">PMID:16942022</ref>. | ||
| + | |||
| + | ====Galantamine derivative (compound 3)==== | ||
| + | Described in the page '[[AChE inhibitors and substrates (Part II)]]' [http://en.wikipedia.org/wiki/Galantamine galanthamine] (<scene name='1w4l/Al/2'>GAL</scene>; <font color='red'><b>colored red</b></font>) is an [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor AChE inhibitor] and it is currently used in therapy of the [http://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's disease] (AD). Conjugate of GAL through the <scene name='1w4l/Al/3'>alkyl linker</scene> (8 carbons, <font color='black'><b>yellow</b></font>) with a <scene name='1w4l/Al/4'>phthalimido moiety</scene> <font color='blueviolet'><b>(blueviolet)</b></font> called '''compound 3''' has a larger affinity for AChE than that of GAL alone. This is similar to previously described cases of bivalent ligands. | ||
| + | A comparison between <scene name='1w4l/Comparison/1'>compound 3</scene>/''Tc''AChE ([[1w4l]]) and <scene name='1w4l/Comparison/2'>galanthamine/TcAChE</scene> structure ([[1dx6]]) shows an identical binding mode of the <font color='red'><b>GAL-moiety (transparent red)</b></font> of '''compound 3''' to that of <font color='blue'><b>GAL alone (blue)</b></font> at the CAS. A <font color='gray'><b>PEG molecule (gray)</b></font> is located at the [http://en.wikipedia.org/wiki/Active_site active site] of the galanthamine/''Tc''AChE structure. The alkyl linker spans the active-site gorge and the phthalimido moiety of '''compound 3''' is situated near Trp279 at the PAS. '''Compound 3''' has higher affinity to ''Tc''AChE than GAL. This can be explained by the higher number of interactions between '''compound 3''' (which interacts not only with residues within CAS but also within PAS) and ''Tc''AChE relative to GAL <ref name="Greenblatt">PMID:15563167</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====CPT-11==== | ||
| + | The drug <scene name='1u65/Cpt_11/1'>CPT-11</scene> <font color='black'><b>(yellow)</b></font> interacts with 13 residues of the <scene name='1u65/Binding_site/1'>active-site gorge</scene> from Trp84 at the bottom to Phe284 at the top ([[1u65]]). Nine of these residues are <scene name='1u65/Binding_site/2'>aromatic</scene> <font color='darkmagenta'><b>(Tyr70, Trp84, Tyr121, Trp279, Phe284, Phe330, Phe331, Tyr334, and His440; colored dark magenta)</b></font>. The contacts made by the drug at the bottom of the gorge involves <scene name='1u65/Binding_site/6'>complementary surface contacts</scene> with Trp84, Tyr121, Phe331, and His440 and, especially, a [http://en.wikipedia.org/wiki/Stacking_(chemistry) stacking interaction] with Phe330. The [http://en.wikipedia.org/wiki/Carbamate carbamate] moiety of [http://en.wikipedia.org/wiki/Irinotecan CPT-11] is seen near residues <scene name='1u65/Binding_site/4'>Phe331 and Tyr334</scene>. <font color='magenta'><b>Carbon C9 (shown in magenta)</b></font> of the carbamate linkage in CPT-11, is 9.3 Å from <scene name='1u65/Binding_site/5'>Ser200</scene> <font color='red'><b>Oγ, the nucleophilic atom </b></font> within the three catalytic residues Ser200, His440, and Glu327. The steric clashes between CPT-11 and ''Tc''AChE residues bar the positioning of CPT-11 near Ser200 Oγ (where [http://en.wikipedia.org/wiki/Hydrolysis hydrolysis] could occur), therefore, ''Tc''AChE can not hydrolyze CPT-11 <ref name="Harel">PMID:15772291</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====BW284C51==== | ||
| + | In a similar fashion to other AChE bivalent inhibitors, <font color='magenta'><b>BW284C51 (BW)</b></font> binds to ''Tc''AChE ([[1e3q]]) at both subsites of its <scene name='1e3q/Active_site/1'>active site</scene> - CAS and PAS. At the CAS, the BW makes a cation-aromatic interaction via its quaternary group to <scene name='1e3q/Active_site/2'>Trp84</scene> <font color='orange'><b>(colored orange)</b></font>. The BW phenyl ring forms an aromatic-aromatic interaction with His440. There is also an electrostatic interaction between the BW proximal quaternary group and Glu199. Near the PAS, BW via its distal quaternary group, interacts with <scene name='1e3q/Active_site/3'>Trp279</scene> <font color='cyan'><b>(colored cyan)</b></font> and forms an aromatic interaction with Tyr334. BW forms hydrogen bond with Tyr121 OH, and makes alkyl interactions with Phe331. The superposition of BW with two other AChE bivalent inhibitors <scene name='1e3q/Active_site/4'>DECA</scene> <font color='gray'><b>(decamethonium, colored gray, [[1acl]])</b></font> and <scene name='1e3q/Active_site/5'>E2020</scene> <font color='blueviolet'><b>(Aricept, colored blueviolet, [[1eve]])</b></font> at the ''Tc''AChE active site gorge reveals similar mode of binding. These 3 inhibitors form [http://en.wikipedia.org/wiki/Cation-pi_interaction cation-π] and π-π interactions with active-site gorge aromatic residues <scene name='1e3q/Active_site/6'>(Tyr70, Trp84, Trp279 and Phe330 or Tyr334)</scene> <font color='black'><b>(colored yellow)</b></font>. The superposition of <scene name='1e3q/Active_site/7'>DECA and E2020</scene> reveals their similar trajectory along the active site gorge, but <scene name='1e3q/Active_site/8'>BW</scene> has a different one. This results in <scene name='1e3q/Active_site/9'>different conformation of Phe330</scene>, which interacts with BW more strongly than with DECA and E2020. The conformations of the other important residues at the active site are similar in all these inhibitor complexes. | ||
| + | It has been shown experimentally that BW and E2020 bind to ''Tc''AChE approximately 100-fold stronger than DECA. These findings have several explanations: ''i)'' E2020 and BW are less flexible than DECA; ''ii)'' the aromatic groups of E2020 and BW form favourable π-π interactions with ''Tc''AChE aromatic residues, in contrast to DECA; and ''iii)'' <scene name='1e3q/Shape/3'>BW</scene> and <scene name='1e3q/Shape/2'>E2020</scene> have aromatic groups and, therefore, occupy more volume and better fit the active-site gorge, than <scene name='1e3q/Shape/4'>string-shaped DECA</scene>. Mutations at the mouse or chicken AChE residues, corresponding to the ''Tc''AChE <scene name='1e3q/Active_site/10'>Tyr70, Trp84, Trp279 and Tyr121</scene> <font color='red'><b>(colored red)</b></font>, cause significant increase of inhibition constant values for all these 3 inhibitors, supporting the notion that these residues are critical for inhibitor-AChE binding <ref name="Felder">PMID:12351819</ref> <ref name="Schalk">PMID:8415649</ref> <ref name="Kryger">PMID:10368299</ref>. | ||
| + | |||
| + | {{Clear}} | ||
| + | |||
| + | ====PEG-SH-350==== | ||
| + | <scene name='1jjb/Active_site/1'>PEG-SH-350</scene> is an untypical acetylcholinesterase inhibitor ([[1jjb]]). It consists of [http://en.wikipedia.org/wiki/Heptamer heptameric] [http://en.wikipedia.org/wiki/Polyethylene_glycol polyethylene glycol] (PEG) with a [http://en.wikipedia.org/wiki/Thiol thiol group] (SH) at the terminus. The thiol group binds close to the <scene name='1jjb/Active_site/4'>catalytic anionic site (CAS)</scene> and the second terminus binds to the <scene name='1jjb/Active_site/5'>peripheral anionic site (PAS)</scene>. PEG-SH-350 interacts with ''Torpedo californica'' acetylcholinesterase via a system of <scene name='1jjb/Active_site/6'>water molecules</scene> <font color='red'><b>(represented by oxygens colored red)</b></font>. Two out of the seven PEG-SH-350 [http://en.wikipedia.org/wiki/Ethylene_glycol ethylene glycol] units are in [http://en.wikipedia.org/wiki/Alkane_stereochemistry ''trans''] <scene name='1jjb/Active_site/7'>conformation</scene> <font color='blue'><b>(colored blue)</b></font>, while the others are in [http://en.wikipedia.org/wiki/Alkane_stereochemistry ''±gauche''] <scene name='1jjb/Active_site/8'>conformation</scene> <ref name="Koellner">PMID:12095250</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====Aricept==== | ||
| + | [http://en.wikipedia.org/wiki/Donepezil Aricept]. Among the most interesting drugs that have been designed to inhibit | ||
| + | [[acetylcholinesterase]] are those that have two binding sites that bind both the peripheral and catatylic sites simultaneously. Such drugs bind strongly and with high specificly. A good example is <scene name='Acetylcholinesterase/1eve_e2020/1'>the E2020/''Tc''AChE (Aricept) complex</scene> ([[1eve]]). It appears that the principal interaction between the aceylcholine and the enzyme is the relatively newly discovered cation-pi interaction between the cationic moiety of the substrate and the many aromatic residues lining the catalytic gorge. Unlike most | ||
| + | interatomic interactions in chemistry, cation-pi interactions are unusual in that their energy hardly changes as the cationic and aromatic ring centers distance vary between 4 and 7 Å, and for a wide variety of relative orientations of the aromatic rings. This gives the substrate an energetically smooth ride down the gorge with few bumps or barriers to impede passage down the gorge. Most acetylcholinesterases have a net negative charge and a large patch of negative potential around the entrance to the active site gorge. This may be useful to attract the positively charged acetycholine substrate to the site. As one travels down the gorge, this potential becomes increasingly more and more negative, reaching a peak at the active site at the base. Because of this potential, the peripherial site is thought to act like a substrate trap, that forces practically every molecule of substrate that reaches the peripheral site to travel down the gorge to the active site. This probably contributes greatly to the extremely rapid rate of degrading the substrate. This whole enzyme therefore acts like a brilliantly designed natural vacuum cleaner that clears the neurotransmitter out of the synapse extremely quickly. Yet to be solved, however, is how the products clear the active site rapidly, whether back through the gorge, or out a back door on the other side of the protein that quickly opens each catalytic cycle (Trp84 | ||
| + | is actually near the surface at the 'underside' of the protein). The X-ray structure of the E2020-''Tc''AChE complex shows that E2020 has a <scene name='1eve/E2020_close_up_with_84_279/13'>unique orientation</scene> along the active-site gorge, extending from the anionic subsite (<scene name='1eve/E2020_close_up_with_84lbld/7'>W84</scene>) of the active site, at the bottom, to the peripheral anionic site (<scene name='1eve/E2020_close_up_with_84_279lbld/5'>near W279</scene>), at the top, via aromatic stacking interactions with conserved aromatic acid residues. E2020 does not, however, interact directly with either the catalytic triad or the 'oxyanion hole' but only <scene name='1eve/E20_interactionshown/8'>indirectly via solvent molecules</scene> <ref name="Kryger"/>. | ||
| + | |||
| + | {{Clear}} | ||
| + | |||
| + | ====Decamethonium==== | ||
| + | Binding sites of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] ([http://www.expasy.org/enzyme/3.1.1.7 EC 3.1.1.7]) with the bisquaternary [http://en.wikipedia.org/wiki/Ligand_(biochemistry) ligand] [http://en.wikipedia.org/wiki/Decamethonium decamethonium] (DME, [[1acl]]). DME is oriented along the <scene name='1acl/Active_site/1'>narrow gorge leading to the active site</scene>; one quaternary group is apposed to <font color='orange'><b>the indole moiety of</b></font> <scene name='1acl/Active_site/2'>W84</scene> (catalytic anionic site, CAS) and the other to <font color='cyan'><b>the indole moiety</b></font> [http://en.wikipedia.org/wiki/Indole] of <scene name='1acl/Active_site/3'>W279</scene>, near the top of the gorge, i.e. the "peripheral" anionic site (PAS). The only major conformational change in the structure of ''Tc''AChE is in the orientation of <scene name='1acl/Active_site/5'>F330</scene> <font color='purple'><b> (purple)</b></font> which lies parallel to the surface of the gorge, near the CAS of ''Tc''AChE which contains the <scene name='1acl/Active_site/4'>catalytic triad</scene> S200, E327 & H440<font color='magenta'><b> (magenta) </b></font>.<ref name="Schalk">PMID:8415649</ref> | ||
| + | </StructureSection> | ||
===Additional Resources=== | ===Additional Resources=== | ||
| - | For additional information, see: [[Alzheimer's Disease]] | + | For additional information, see: |
| + | *[[Alzheimer's Disease]] | ||
| + | *[[Acetylcholinesterase inhibitors]] | ||
==References== | ==References== | ||
| Line 98: | Line 158: | ||
[[Category: AChE inhibitors]] | [[Category: AChE inhibitors]] | ||
[[Category: inhibitor]] | [[Category: inhibitor]] | ||
| - | [[Category: cholinesterases]] | ||
[[Category: acetylcholine]] | [[Category: acetylcholine]] | ||
[[Category: cation-pi]] | [[Category: cation-pi]] | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see:
References
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Botti SA, Felder CE, Lifson S, Sussman JL, Silman I. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J. 1999 Nov;77(5):2430-50. PMID:10545346
- ↑ 3.0 3.1 3.2 Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP, Sussman JL. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A. Nat Struct Biol. 1997 Jan;4(1):57-63. PMID:8989325
- ↑ Sanson B, Nachon F, Colletier JP, Froment MT, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, Weik M. Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime (dagger). J Med Chem. 2009 Jul 30. PMID:19642642 doi:10.1021/jm900433t
- ↑ 5.0 5.1 5.2 Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL. Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry. 1999 Jun 1;38(22):7032-9. PMID:10353814 doi:http://dx.doi.org/10.1021/bi982678l
- ↑ 6.0 6.1 6.2 Kryger G, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with E2020 (Aricept): implications for the design of new anti-Alzheimer drugs. Structure. 1999 Mar 15;7(3):297-307. PMID:10368299
- ↑ 7.0 7.1 7.2 Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9031-5. PMID:8415649
- ↑ Dvir H, Wong DM, Harel M, Barril X, Orozco M, Luque FJ, Munoz-Torrero D, Camps P, Rosenberry TL, Silman I, Sussman JL. 3D structure of Torpedo californica acetylcholinesterase complexed with huprine X at 2.1 A resolution: kinetic and molecular dynamic correlates. Biochemistry. 2002 Mar 5;41(9):2970-81. PMID:11863435
- ↑ 9.0 9.1 9.2 Greenblatt HM, Kryger G, Lewis T, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with (-)-galanthamine at 2.3 A resolution. FEBS Lett. 1999 Dec 17;463(3):321-6. PMID:10606746
- ↑ 10.0 10.1 Ravelli RB, Raves ML, Ren Z, Bourgeois D, Roth M, Kroon J, Silman I, Sussman JL. Static Laue diffraction studies on acetylcholinesterase. Acta Crystallogr D Biol Crystallogr. 1998 Nov 1;54(Pt 6 Pt 2):1359-66. PMID:10089512
- ↑ Bar-On P, Millard CB, Harel M, Dvir H, Enz A, Sussman JL, Silman I. Kinetic and structural studies on the interaction of cholinesterases with the anti-Alzheimer drug rivastigmine. Biochemistry. 2002 Mar 19;41(11):3555-64. PMID:11888271
- ↑ Harel M, Sonoda LK, Silman I, Sussman JL, Rosenberry TL. Crystal structure of thioflavin T bound to the peripheral site of Torpedo californica acetylcholinesterase reveals how thioflavin T acts as a sensitive fluorescent reporter of ligand binding to the acylation site. J Am Chem Soc. 2008 Jun 25;130(25):7856-61. Epub 2008 May 31. PMID:18512913 doi:http://dx.doi.org/10.1021/ja7109822
- ↑ Paz A, Roth E, Ashani Y, Xu Y, Shnyrov VL, Sussman JL, Silman I, Weiner L. Structural and functional characterization of the interaction of the photosensitizing probe methylene blue with Torpedo californica acetylcholinesterase. Protein Sci. 2012 Jun 1. doi: 10.1002/pro.2101. PMID:22674800 doi:10.1002/pro.2101
- ↑ Dym O, Song W, Felder C, Roth E, Shnyrov V, Ashani Y, Xu Y, Joosten RP, Weiner L, Sussman JL, Silman I. The Impact of Crystallization Conditions on Structure-Based Drug Design: A Case Study on the Methylene Blue/Acetylcholinesterase Complex. Protein Sci. 2016 Mar 14. doi: 10.1002/pro.2923. PMID:26990888 doi:http://dx.doi.org/10.1002/pro.2923
- ↑ Colletier JP, Bourgeois D, Sanson B, Fournier D, Sussman JL, Silman I, Weik M. Shoot-and-Trap: use of specific x-ray damage to study structural protein dynamics by temperature-controlled cryo-crystallography. Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):11742-7. Epub 2008 Aug 13. PMID:18701720
- ↑ 16.0 16.1 Haviv H, Wong DM, Greenblatt HM, Carlier PR, Pang YP, Silman I, Sussman JL. Crystal packing mediates enantioselective ligand recognition at the peripheral site of acetylcholinesterase. J Am Chem Soc. 2005 Aug 10;127(31):11029-36. PMID:16076210 doi:http://dx.doi.org/10.1021/ja051765f
- ↑ Wong DM, Greenblatt HM, Dvir H, Carlier PR, Han YF, Pang YP, Silman I, Sussman JL. Acetylcholinesterase complexed with bivalent ligands related to huperzine a: experimental evidence for species-dependent protein-ligand complementarity. J Am Chem Soc. 2003 Jan 15;125(2):363-73. PMID:12517147 doi:http://dx.doi.org/10.1021/ja021111w
- ↑ Rydberg EH, Brumshtein B, Greenblatt HM, Wong DM, Shaya D, Williams LD, Carlier PR, Pang YP, Silman I, Sussman JL. Complexes of alkylene-linked tacrine dimers with Torpedo californica acetylcholinesterase: Binding of Bis5-tacrine produces a dramatic rearrangement in the active-site gorge. J Med Chem. 2006 Sep 7;49(18):5491-500. PMID:16942022 doi:http://dx.doi.org/10.1021/jm060164b
- ↑ Felder CE, Harel M, Silman I, Sussman JL. Structure of a complex of the potent and specific inhibitor BW284C51 with Torpedo californica acetylcholinesterase. Acta Crystallogr D Biol Crystallogr. 2002 Oct;58(Pt 10 Pt 2):1765-71. Epub, 2002 Sep 28. PMID:12351819
- ↑ 20.0 20.1 Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9031-5. PMID:8415649
- ↑ Koellner G, Steiner T, Millard CB, Silman I, Sussman JL. A neutral molecule in a cation-binding site: specific binding of a PEG-SH to acetylcholinesterase from Torpedo californica. J Mol Biol. 2002 Jul 19;320(4):721-5. PMID:12095250
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Michal Harel, Jaime Prilusky, David Canner