Prion
From Proteopedia
(Difference between revisions)
| (36 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
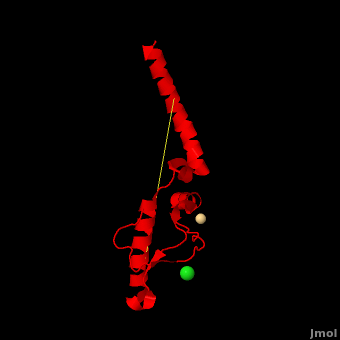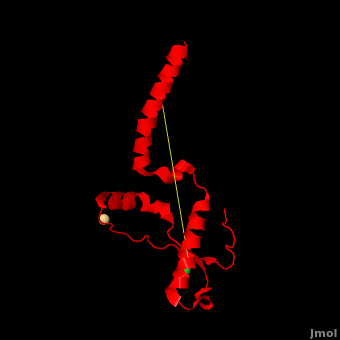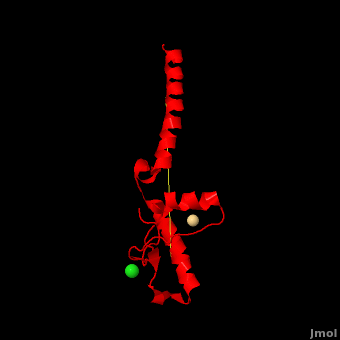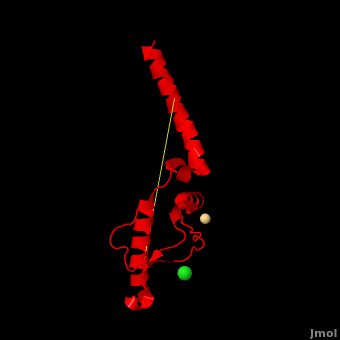
| - | + | <StructureSection load='' size='350' side='right' scene='Prion/Cv/3' caption='Human major prion protein complex with Cd+ (pink) and Cl- (green) ions (PDB code [[3haf]])'> | |
| - | + | ||
| - | + | ||
| + | {{Clear}} | ||
| + | __TOC__ | ||
| + | ==Function== | ||
| + | '''Prion''' (PrP) is a protein which becomes infectious upon undergoing conformation change to an amyloid form, which is self-propagating and becomes resistant to protease degradation. The fungus ''Podospora anserine'' has a prion-like protein HET-S which undergoes a conformation change to amyloid form which prevents its colony from merging with non-compatible colonies. Yeast prion proteins are Sup35 and Ure2. For more details see<br /> | ||
| + | *[[Prion protein]]<br /> | ||
| + | *[[Human Prion Protein Dimer]]<br /> | ||
| + | *[[Doppel]]<br /> | ||
| + | *[[Group:SMART:A Physical Model of the Structure of GNNQQNY from Yeast Prion Sup35]]<br /> | ||
| + | Click here to see <scene name='Prion/Cv/2'>example of prion unfolding</scene> (morph was taken from [http://molmovdb.org/cgi-bin/movie.cgi Gallery of Morphs] of the [http://molmovdb.org Yale Morph Server]). | ||
| + | == Dominant-negative Effects in Prion Diseases== | ||
| - | + | Insights from Molecular Dynamics Simulations on <scene name='Journal:JBSD:4/Cv/2'>Mouse Prion Protein Chimeras</scene> <ref>doi 10.1080/07391102.2012.712477</ref>. | |
| - | + | The key event in prion diseases is the conformational conversion from the cellular form of the [[Prion_protein|prion protein]] (PrP<sup>C</sup>) to its pathogenic scrapie form PrP<sup>Sc</sup> (or prion). PrP<sup>Sc</sup> is the sole causative agent of prion diseases which self-propagates by converting PrP<sup>C</sup> to nascent PrP<sup>Sc</sup>. Mutations in the open reading sequence of the [[Prion_protein|prion protein]] gene can introduce changes in the protein structure and alter PrP<sup>Sc</sup> formation and propagation, possibly by (de)stabilizing the physiological folding of PrP<sup>C</sup> and/or affecting its interactions with some yet unknown cellular factors. Some PrP polymorphisms may even inhibit the wild-type (WT) PrP<sup>C</sup> from being converted to PrP<sup>Sc</sup>, with the so-called “dominant-negative” effect. | |
| - | + | Here we use molecular dynamics simulations to investigate the structural determinants of the globular domain in engineered Mouse (Mo) PrP variants, in WT human (Hu) PrP (PDB: [[1hjn]]) and in WT MoPrP (PDB: [[1xyx]]). The Mo PrP variants investigated here contain one or two residues from ''Homo sapiens'' and are denoted “MoPrP chimeras”. <scene name='Journal:JBSD:4/Cv/3'>Some of them are resistant to PrP<sup>Sc</sup> infection</scene> <span style="color:yellow;background-color:black;font-weight:bold;">(colored in yellow)</span> in ''in vivo'' or in ''in vitro'' cell-culture experiments, the <scene name='Journal:JBSD:4/Cv/7'>others are not</scene> <font color='darkmagenta'><b>(in darkmagenta)</b></font>. Our main results are the following: (i) The chimeras resistant to PrP<sup>Sc</sup> infection show <scene name='Journal:JBSD:4/Cv/8'>shorter intramolecular distances</scene> between the α1 helix and N-terminal of α3 helix than HuPrP, MoPrP and the non-resistant chimeras (<scene name='Journal:JBSD:4/Cv/12'>click here to see morph</scene>). This is due to stronger specific interactions between these two regions, mainly the <scene name='Journal:JBSD:4/Cv/9'>Y149-D202 and D202-Y157 (in Hu numbering and hereafter) hydrogen bonds</scene> and the <scene name='Journal:JBSD:4/Cv/10'>R156-E196 salt bridge</scene>. (ii) The β2-α2 <scene name='Journal:JBSD:4/Cv1/2'>loop (residues 167-171)</scene> of PrP<sup>C</sup> is known to differ in its conformation across different species and is suggested to be responsible for the species barrier of PrP<sup>Sc</sup> propagation. Our simulations detect exchanges between different conformations in this loop which can be categorized into two distinct patterns: some chimeras experience a 3<sub>10</sub>-helix/turn pattern like in MoPrP and others show a bend/turn pattern like in HuPrP. In the <span style="color:lime;background-color:black;font-weight:bold;">Mo-like pattern (colored in green)</span>, 3<sub>10</sub>-helix conformation is stabilized by the <scene name='Journal:JBSD:4/Cv1/3'>Q168-P165 and Y169-V166 hydrogen bonds</scene>. In the <font color='darkred'><b>Hu-like pattern (colored in darkred)</b></font>, a <scene name='Journal:JBSD:4/Cv1/4'>D167-S170 hydrogen bond</scene> stabilizes the bend conformation. Interestingly, the dominant-negative effect of MoPrP chimeras over WT MoPrP occurs if the chimera not only resists PrP<sup>Sc</sup> infection but also adopts the Mo-like pattern of exchanges between conformations in the β2-α2 loop. This suggests that the compatible loop conformation allows these dominant-negative chimeras to interfere with the conversion of MoPrP to PrP<sup>Sc</sup>. | |
| - | + | The structural features presented here indicate that stronger interactions between α1 helix and N-terminal of α3 helix are related to the resistance to PrP<sup>C</sup> → PrP<sup>Sc</sup> conversion, while the β2-α2 loop conformation may play an important role in the dominant-negative effect. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==3D structures of prion== | ==3D structures of prion== | ||
| + | [[Prion 3D structures ]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
- ↑ Cong X, Bongarzone S, Giachin G, Rossetti G, Carloni P, Legname G. Dominant-negative effects in prion diseases: insights from molecular dynamics simulations on mouse prion protein chimeras. J Biomol Struct Dyn. 2012 Aug 30. PMID:22934595 doi:10.1080/07391102.2012.712477