Methionine adenosyltransferase
From Proteopedia
| Line 14: | Line 14: | ||
== Structure == | == Structure == | ||
<StructureSection load='5a1i' size='400' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='5a1i' size='400' side='right' caption='Caption for this structure' scene=''> | ||
| - | MAT consists of α and β subunits. The MATα1 and <scene name='90/907472/Mat2a/4'>MATα2</scene> subunits are catalytic subunits while MATβ is a regulatory subunit. Not much is currently known about the function of this regulatory subunit and how it regulates the function of the enzyme. The subunits are encoded on different genes in humans, so they are created separately and can then come together to form various complexes, such as MATαβ or MATα2 dimers <ref name="Murray et al." />. | + | MAT consists of α and β subunits. The MATα1 and <scene name='90/907472/Mat2a/4'>MATα2</scene> subunits are catalytic subunits while MATβ is a regulatory subunit. Not much is currently known about the function of this regulatory subunit and how it regulates the function of the enzyme<ref>DOI:10.1107/S2052252514012585</ref>. The subunits are encoded on different genes in humans, so they are created separately and can then come together to form various complexes, such as MATαβ or MATα2 dimers <ref name="Murray et al." />. |
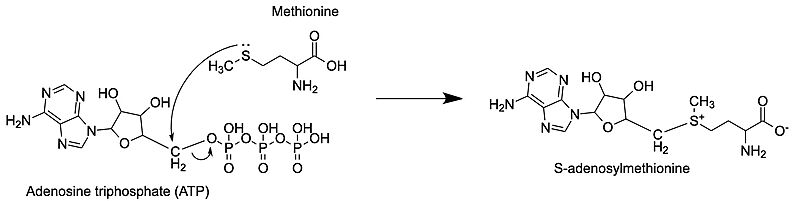
The <scene name='90/907472/Substrates/1'>substrates</scene> used by the enzyme are methionine and ATP. Notably, ATP is not used as a source of energy in this reaction like it is for many other processes. Instead, it is used as a substrate in the synthesis reaction. Methionine and ATP enter the active site and are stabilized by residues present there, including lysine and histidine. Once the reaction begins to take place, methionine flips toward the 5' carbon of the adenosine sugar. Following nucleophilic attack of the sulfur on the carbon, the C-O bond between the phosphates and the carbon breaks, and the <scene name='90/907472/Product/2'>products</scene> are formed (tripolyphosphate not pictured). SAM is released from the active site first. MAT also catalyzes hydrolysis of the tripolyphosphate into pyrophosphate and orthophosphate, which are then released from the active site <ref>Niland CN, Ghosh A, Cahill SM, Schramm VL. Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A. ACS Biochemistry. 2021 Mar 3;60 (10) 791-801. doi: https://doi.org/10.1021/acs.biochem.0c00998</ref>. The biological assembly of rat S-adenosylmethionine synthetase is <scene name='49/493038/Cv/7'>homotetramer</scene>. The <scene name='49/493038/Cv/8'>active site of MAT is located between adjacent subunuts</scene><ref>PMID:12888348</ref>. | The <scene name='90/907472/Substrates/1'>substrates</scene> used by the enzyme are methionine and ATP. Notably, ATP is not used as a source of energy in this reaction like it is for many other processes. Instead, it is used as a substrate in the synthesis reaction. Methionine and ATP enter the active site and are stabilized by residues present there, including lysine and histidine. Once the reaction begins to take place, methionine flips toward the 5' carbon of the adenosine sugar. Following nucleophilic attack of the sulfur on the carbon, the C-O bond between the phosphates and the carbon breaks, and the <scene name='90/907472/Product/2'>products</scene> are formed (tripolyphosphate not pictured). SAM is released from the active site first. MAT also catalyzes hydrolysis of the tripolyphosphate into pyrophosphate and orthophosphate, which are then released from the active site <ref>Niland CN, Ghosh A, Cahill SM, Schramm VL. Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A. ACS Biochemistry. 2021 Mar 3;60 (10) 791-801. doi: https://doi.org/10.1021/acs.biochem.0c00998</ref>. The biological assembly of rat S-adenosylmethionine synthetase is <scene name='49/493038/Cv/7'>homotetramer</scene>. The <scene name='49/493038/Cv/8'>active site of MAT is located between adjacent subunuts</scene><ref>PMID:12888348</ref>. | ||
Revision as of 11:11, 17 April 2022
Methionine adenosyltransferase (MAT) or S-adenosylmethionine synthetase (SAM synthetase) synthesizes S-adenosylmethionine (SAM or AdoMet) from the substrates adenosine triphosphate (ATP) and methionine. ATP isn’t used as a source of energy like it is in other reactions but gets a methionine added onto the 5th carbon while the three phosphate groups are broken down and released from the active site. This enzyme is conserved and found in many organisms, so it is essential for life. Problems with this enzyme have been shown to cause diseases such as various cancers.
Contents |
Relevance
The product of this enzymatic reaction, SAM, is the universal methyl donor of metabolism. SAM is involved in N-methylation, O-methylation and C-methylation, yielding S-adenosyl homocysteine as a product that gets recycled by the one-carbon metabolism. Radical SAM enzymes break down SAM into an adenosyl radical and methionine, enabling a host of otherwise difficult to achieve reactions, e.g. in molybdenum cofactors biosynthesis[1]. Accumulation of S-adenosyl homocysteine (or homocysteine itself) indicates an imbalance in supply and demand for SAM in the organism. Methionine metabolism impairment in liver diseases is related in alteration in MAT[2].
Function and reaction mechanism
S-adenosylmethionine synthetase or S-adenosylmethionine synthase or S-adenosylmethionine transferase or methionine adenosyltransferase (MAT) catalyzes the conversion of methionine and ATP to S-adenosylmethionine (AdoMet), pyrophosphate (PPi) and orthophosphate (Pi). The catalytic entity of MAT is a dimer. MAT cofactors are Mg+2 (or Co+2) and K+ ions[3].
The nucleophilic sulfur atom of methionine attacks the slightly positive 5' carbon of the adenosine sugar unit. Following this, the bond from the 5' carbon to the oxygen breaks, separating the tripolyphosphate from the newly formed S-adenosylmethionine (SAM) [4]. This is an example of an SN2 reaction, where the substrates move through a transition state to then form the products. The products are only released after the methionine binds and the C-O bond breaks.
Structure
| |||||||||||
3D structures of S-adenosylmethionine synthetase
S-adenosylmethionine synthetase 3D structures
17-April-2022
References
- ↑ Hanzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci U S A. 2004 Aug 31;101(35):12870-5. Epub 2004 Aug 18. PMID:15317939 doi:10.1073/pnas.0404624101
- ↑ Mato JM, Alvarez L, Ortiz P, Mingorance J, Duran C, Pajares MA. S-adenosyl-L-methionine synthetase and methionine metabolism deficiencies in cirrhosis. Adv Exp Med Biol. 1994;368:113-7. PMID:7741002
- ↑ Takusagawa F, Kamitori S, Markham GD. Structure and function of S-adenosylmethionine synthetase: crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28 angstroms resolution. Biochemistry. 1996 Feb 27;35(8):2586-96. PMID:8611562 doi:http://dx.doi.org/10.1021/bi952604z
- ↑ 4.0 4.1 4.2 4.3 Murray B, Antonyuk SV, Marina A, Lu SC, Mato JM, Hasnain SS, Rojas Al. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. PNAS. 2016 Feb 8;113 (8) 2104-2109. doi: https://doi.org/10.1073/pnas.1510959113
- ↑ Murray B, Antonyuk SV, Marina A, Van Liempd SM, Lu SC, Mato JM, Hasnain SS, Rojas AL. Structure and function study of the complex that synthesizes S-adenosylmethionine. IUCrJ. 2014 Jun 12;1(Pt 4):240-9. doi: 10.1107/S2052252514012585. eCollection, 2014 Jul 1. PMID:25075345 doi:http://dx.doi.org/10.1107/S2052252514012585
- ↑ Niland CN, Ghosh A, Cahill SM, Schramm VL. Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A. ACS Biochemistry. 2021 Mar 3;60 (10) 791-801. doi: https://doi.org/10.1021/acs.biochem.0c00998
- ↑ Gonzalez B, Pajares MA, Hermoso JA, Guillerm D, Guillerm G, Sanz-Aparicio J. Crystal structures of methionine adenosyltransferase complexed with substrates and products reveal the methionine-ATP recognition and give insights into the catalytic mechanism. J Mol Biol. 2003 Aug 8;331(2):407-16. PMID:12888348
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Anna Postnikova, Michal Harel, Kia Yang, Michael O'Shaughnessy, Alexander Berchansky, Jaime Prilusky