Telomerase
From Proteopedia
(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
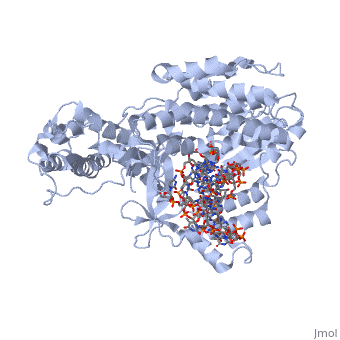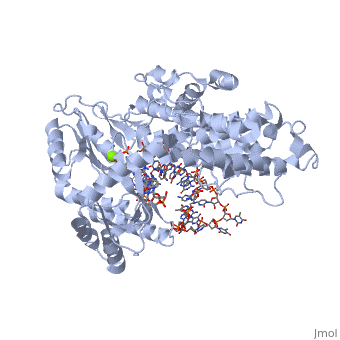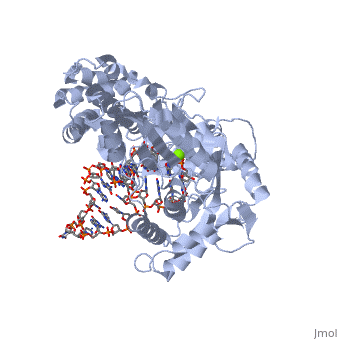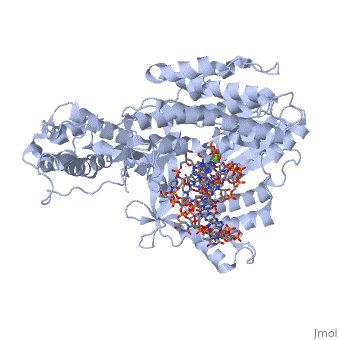
<StructureSection load='3kyl' size='350' side='right' caption='Telomerase: bound to telomeric DNA complex with Ca+2 ion (green) [[3kyl]]' scene=''> | <StructureSection load='3kyl' size='350' side='right' caption='Telomerase: bound to telomeric DNA complex with Ca+2 ion (green) [[3kyl]]' scene=''> | ||
== Introduction == | == Introduction == | ||
| - | The telomerase is a complex composed of | + | The '''telomerase''' is a complex composed of a '''Telomerase RNA''' primer and protein [[Telomerase Reverse Transcriptase]] that adds G-rich repeated DNA sequences to a 3' DNA strand in eukaryotic cells <ref name='complex'>DOI: 10.1146/annurev.bi.61.070192.000553</ref>. This ribonucleoprotein acts as a reverse transcriptase that creates a DNA sequence complementary to its RNA primer in its catalytic subunit, TERT <ref name='end'>DOI: 10.1002/anie.201002387</ref>. The sequences attached to the end of DNA is non-coding DNA that functions as protection from mutation and degradation. As DNA replications occurs, the polymerase that copies in the 5' to 3' direction, cannot copy the end of the lagging strand which leaves unpaired bases that may code for a specific gene <ref name='corey'>DOI 10.1016/j.chembiol.2009.12.001</ref>. Multiple unpaired ends can form hydrogen bonds or even swap genetic material. The telomerase fixes this issue and lengthens the strands, because the replication process slowly shortens the ends of chromosomes. This enzyme also plays a role in coordinating cell division. Complementary issues of aging and cancer describe the inactivation or over-activation of telomerase activity <ref name='corey'/>. |
== History == | == History == | ||
| Line 14: | Line 14: | ||
'''Overall Structure''' | '''Overall Structure''' | ||
| - | <scene name='60/602706/Telomerase/1'>Telomerase</scene> (protein in | + | <scene name='60/602706/Telomerase/1'>Telomerase</scene> (protein in grey; RNA in green; DNA in red) acts as both a monomer and dimer. The monomer refers to the overall protein and its catalytic subunit TERT, made up of an amino acid polymer, containing approximately 5,000 atoms. This protein binds with the RNA template, <scene name='60/602706/Ter/1'>TER</scene>, that TERT uses to add DNA to form a dimer-like structure <ref name='TER'>doi:10.1038/nature07283</ref>. The RNA has a molecular size between 200 and 500 kDA, depending on the organism <ref name='complex'/>. Both the protein and RNA components are highly conserved structures among phylogenetic groups. <scene name='60/602706/Tert/2'>TERT</scene> is organized into a ring-like structure that shares common features with other reverse transcriptases (in viruses for example) and DNA polymerases. The RNA-DNA heteroduplex lies in the interior of the ring and positions the 3' end of the DNA primer at the active site to the telomerse can be enlongated. The substrate binding within the ring can accomodate 7 to 8 bases of double-stranded nucleic acid <ref name='TER'/>. |
'''Architecture of TERT Structure''' | '''Architecture of TERT Structure''' | ||
| Line 27: | Line 27: | ||
The reverse transcription process begins by the telomerase RNA binding to the 3' end of the overhanging DNA sequence. Once primed, the G-rich sequence (~6-8 nucleotides) polymerizes the end of the DNA. The RNA primer then translocates down the new DNA sequence and polymerizes again. This process repeats until the entire telomere is formed. After one side of the telomere is formed, the telomerase attaches the complementary nucleotide bases to complete the double-stranded telomere at the end of the DNA double helix. | The reverse transcription process begins by the telomerase RNA binding to the 3' end of the overhanging DNA sequence. Once primed, the G-rich sequence (~6-8 nucleotides) polymerizes the end of the DNA. The RNA primer then translocates down the new DNA sequence and polymerizes again. This process repeats until the entire telomere is formed. After one side of the telomere is formed, the telomerase attaches the complementary nucleotide bases to complete the double-stranded telomere at the end of the DNA double helix. | ||
| - | |||
==Telomerase 3D structures== | ==Telomerase 3D structures== | ||
| - | + | [[Telomerase 3D structures]] | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | + | [[Category:Topic Page]] | |
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 Blackburn EH. Telomerases. Annu Rev Biochem. 1992;61:113-29. PMID:1497307 doi:http://dx.doi.org/10.1146/annurev.bi.61.070192.000553
- ↑ 2.0 2.1 Blackburn EH. Telomeres and telomerase: the means to the end (Nobel lecture). Angew Chem Int Ed Engl. 2010 Oct 4;49(41):7405-21. doi: 10.1002/anie.201002387. PMID:20821774 doi:http://dx.doi.org/10.1002/anie.201002387
- ↑ 3.0 3.1 3.2 3.3 3.4 Corey DR. Telomeres and telomerase: from discovery to clinical trials. Chem Biol. 2009 Dec 24;16(12):1219-23. doi: 10.1016/j.chembiol.2009.12.001. PMID:20064431 doi:http://dx.doi.org/10.1016/j.chembiol.2009.12.001
- ↑ 4.0 4.1 4.2 Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008 Oct 2;455(7213):633-7. Epub 2008 Aug 31. PMID:18758444 doi:http://dx.doi.org/10.1038/nature07283
- ↑ 5.0 5.1 Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010 Apr;17(4):513-8. Epub 2010 Mar 28. PMID:20357774 doi:10.1038/nsmb.1777