BASIL2022GV3HDT
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Online Resources (bioinformatic software) == | == Online Resources (bioinformatic software) == | ||
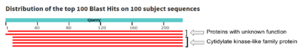
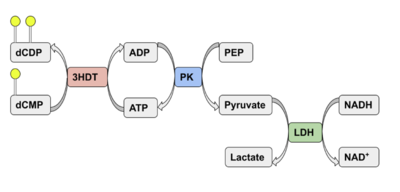
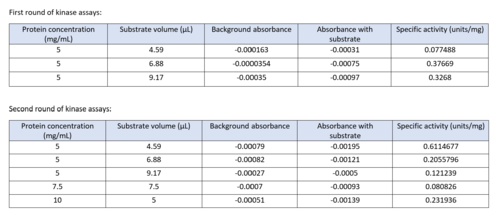
| - | We used a variety of ''in silico'' | + | We used a variety of ''in silico'' tools with our protein, 3HDT to find similarities with other amino acid sequences, protein family matches, and structural comparisons to known proteins in the PDB. Below are the recorded results and information from each database. From this information, a hypothesized function was created for 3HDT and potential substrates were selected such as dCMP. |
===='''BLASTp'''==== | ===='''BLASTp'''==== | ||
Revision as of 02:33, 20 April 2022
Characterizing Putative Kinase 3HDT
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Proteopedia Page Contributors and Editors (what is this?)
Jesse D. Rothfus, Autumn Forrester, Bonnie Hall, Jaime Prilusky