BASIL2022GV3HDT
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== ''In silico'' anaylsis == | == ''In silico'' anaylsis == | ||
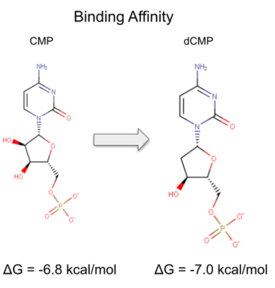
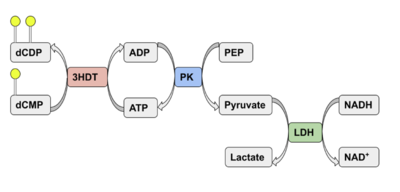
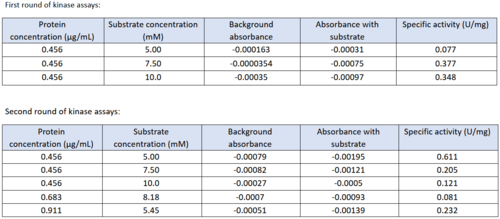
| - | We used a variety of ''in silico'' tools with our protein, 3HDT to find similarities with other amino acid sequences, protein | + | We used a variety of ''in silico'' tools with our protein, 3HDT, to find similarities with other amino acid sequences, protein families, and 3-dimensional structure of known proteins in the PDB. Below are the recorded results and information from each database. From this information, a hypothesized function was created for 3HDT and potential substrates such as dCMP were selected. |
===='''BLASTp'''==== | ===='''BLASTp'''==== | ||
| - | Beginning with BLASTp<ref>National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].</ref>, we queried the FASTA sequence for our protein, PDB ID 3hdt. The alignment of our query sequence with the NK superfamily shows a high degree of overlap indicating this protein is likely a member. The importance of this is that the cytidylate kinase family is a member of the NK superfamily, supporting our claim that 3hdt is a cytidylate kinase. Also, the query hits show | + | Beginning with BLASTp<ref>National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].</ref>, we queried the FASTA sequence for our protein, PDB ID 3hdt. The alignment of our query sequence with the NK superfamily shows a high degree of overlap indicating this protein is likely a member. The importance of this is that the cytidylate kinase family is a member of the NK superfamily, supporting our claim that 3hdt is a cytidylate kinase. Also, the query hits show several cytidylate kinase-like family proteins almost completely aligning with our protein 3HDT. |
[[Image:BLASTp image 1.png |600px| left | thumb | BLASTp Alignment showing a hit with cytidylate kinase-like family, part of the NK superfamily.]] | [[Image:BLASTp image 1.png |600px| left | thumb | BLASTp Alignment showing a hit with cytidylate kinase-like family, part of the NK superfamily.]] | ||
| Line 21: | Line 21: | ||
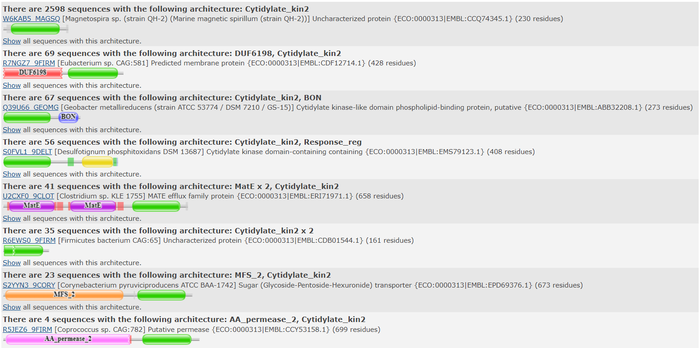
===='''Pfam'''==== | ===='''Pfam'''==== | ||
We used the FASTA sequence from 3HDT to do a comparative search in Pfam<ref>Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman | We used the FASTA sequence from 3HDT to do a comparative search in Pfam<ref>Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman | ||
| - | Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913</ref> to find similar protein families our protein may belong. Pfam predicted that our protein is part of the cytidylate kinase which was also shown in the BLASTp results. | + | Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913</ref> to find similar protein families that our protein may belong to. Pfam predicted that our protein is part of the cytidylate kinase-like family which was also shown in the BLASTp results. |
[[Image:Pfam search resultsJRAF2022.png |800px| center | thumb | Pfam query aligned with a cytidylate kinase-like family.]] | [[Image:Pfam search resultsJRAF2022.png |800px| center | thumb | Pfam query aligned with a cytidylate kinase-like family.]] | ||
Revision as of 17:05, 26 April 2022
Characterizing Putative Kinase 3HDT
| |||||||||||
References
- ↑ National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].
- ↑ Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
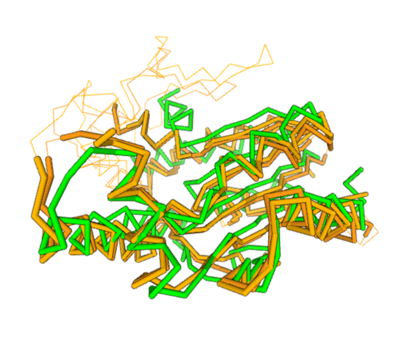
- ↑ Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
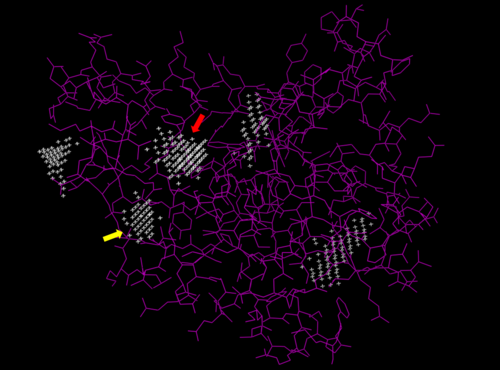
- ↑ J. Yu, Y. Zhou, I. Tanaka, M. Yao, Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics, 26(1), 46-52, (2010) [PMID: 19846440]
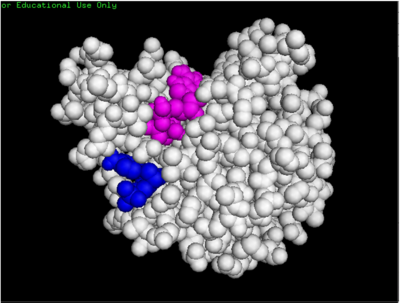
- ↑ Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.
- ↑ The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Jesse D. Rothfus, Autumn Forrester, Bonnie Hall, Jaime Prilusky