NKX2.5 Homeodomain
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | ||
= General Information = | = General Information = | ||
| - | The transcription factor, NKX2.5 is one of many proteins classified as a homeodomain, and functions to regulate structural development in eukaryotes. These proteins share a characteristic evolutionarily conserved fold containing three alpha-helices. <ref> PMID: 7979246 </ref>. DNA-binding is mediated through the insertion of the c-terminal side <scene name='91/911264/Major_groove_interaction/2'>alpha-helix</scene> into the major groove, allowing for base-reside interactions. This allows homeodomains to locate and bind specific DNA sequences, leading to transcriptional activation or repression <ref> PMID: 26464018 </ref>. The homeodomain of NKX2.5 is flanked by both a N and C-terminal regulatory domain. This puts the biological protein at 324 residues with the homeodomain consisting of residues 138-197 <ref> PMID: 22849347 </ref> | + | The transcription factor, NKX2.5 is one of many proteins classified as a homeodomain, and functions to regulate structural development in eukaryotes. These proteins share a characteristic evolutionarily conserved fold containing <scene name='91/911264/Three_alpha_helices/2'>three alpha-helices</scene>. <ref> PMID: 7979246 </ref>. DNA-binding is mediated through the insertion of the c-terminal side <scene name='91/911264/Major_groove_interaction/2'>alpha-helix</scene> into the major groove, allowing for base-reside interactions. This allows homeodomains to locate and bind specific DNA sequences, leading to transcriptional activation or repression <ref> PMID: 26464018 </ref>. The homeodomain of NKX2.5 is flanked by both a N and C-terminal regulatory domain. This puts the biological protein at 324 residues with the homeodomain consisting of residues 138-197 <ref> PMID: 22849347 </ref> |
[[Image:Map.png]] Research into the structure and function of NKX2.5 has mainly been focused on the DNA-binding homeodomain, as mutations in this region have been linked to specific diseases <ref name="Schott"> PMID: 9651244</ref>. This page will focus on the specific interactions of the homeodomain of NKX2.5 with DNA, and how these interactions relate to one of the transcription factor's primary function - heart development. | [[Image:Map.png]] Research into the structure and function of NKX2.5 has mainly been focused on the DNA-binding homeodomain, as mutations in this region have been linked to specific diseases <ref name="Schott"> PMID: 9651244</ref>. This page will focus on the specific interactions of the homeodomain of NKX2.5 with DNA, and how these interactions relate to one of the transcription factor's primary function - heart development. | ||
Revision as of 02:06, 4 May 2022
Contents |
General Information
The transcription factor, NKX2.5 is one of many proteins classified as a homeodomain, and functions to regulate structural development in eukaryotes. These proteins share a characteristic evolutionarily conserved fold containing . [1]. DNA-binding is mediated through the insertion of the c-terminal side into the major groove, allowing for base-reside interactions. This allows homeodomains to locate and bind specific DNA sequences, leading to transcriptional activation or repression [2]. The homeodomain of NKX2.5 is flanked by both a N and C-terminal regulatory domain. This puts the biological protein at 324 residues with the homeodomain consisting of residues 138-197 [3]
 Research into the structure and function of NKX2.5 has mainly been focused on the DNA-binding homeodomain, as mutations in this region have been linked to specific diseases [4]. This page will focus on the specific interactions of the homeodomain of NKX2.5 with DNA, and how these interactions relate to one of the transcription factor's primary function - heart development.
Research into the structure and function of NKX2.5 has mainly been focused on the DNA-binding homeodomain, as mutations in this region have been linked to specific diseases [4]. This page will focus on the specific interactions of the homeodomain of NKX2.5 with DNA, and how these interactions relate to one of the transcription factor's primary function - heart development.
Clinical Relevance
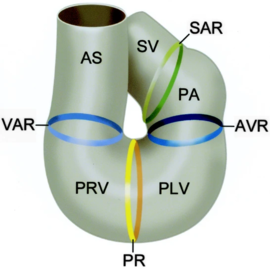
In the case of NKX2.5, the protein works in conjunction with multiple other transcription factors during cardiogenesis [6],[7]. Recently, research has been focused on NKX2.5 as mutations in the DNA binding residues, and structural support residues of the protein have been linked to congenital heart disease. Specifically, NKX2.5 mutations have been linked to etiologies involving both atrial and septal defects, deficient atrioventricular node conduction, and more complex mutations such as Tetralogy of Fallot and Hypoplastic Left Heart Syndrome [8],[4],[9]. These phenotypes are speculated to arise as a result of decreased DNA-binding of NKX2.5 during cardiogenesis. Thi [10]. Specifically, issues take root during cardiac loop formation, which takes place approximately 24 days after embryogenesis. At this time, the single vessel present in the embryo loops back upon itself, allowing the vessel to come in contact with itself. This leads to the formation of a four-chambered, primitive heart with the great vessels (shown in the figure from Gittenberger-de Groot et. al)[5]. Throughout this point in development, NKX2.5 along with other transcription factors are essential in facilitating initial loop formation. Therefore, mutations that affect NKX2.5 function have been tied to the defects mentioned above [11]. Additionally, studies have shown that NKX2.5 continues to function after development by maintaining conductivity through the heart's natural pace-maker system. Although the process by which this occurs is slightly less well known, it does suggest NKX2.5 is important throughout life [12].
Protein / DNA Interactions
Scene
Mutational Analysis
Protein / DNA interactions
Structural highlights
</StructureSection>
References
- ↑ Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487-526. doi: 10.1146/annurev.bi.63.070194.002415. PMID:7979246 doi:http://dx.doi.org/10.1146/annurev.bi.63.070194.002415
- ↑ Burglin TR, Affolter M. Homeodomain proteins: an update. Chromosoma. 2016 Jun;125(3):497-521. doi: 10.1007/s00412-015-0543-8. Epub 2015, Oct 13. PMID:26464018 doi:http://dx.doi.org/10.1007/s00412-015-0543-8
- ↑ Pradhan L, Genis C, Scone P, Weinberg EO, Kasahara H, Nam HJ. Crystal structure of the human NKX2.5 homeodomain in complex with DNA target. Biochemistry. 2012 Aug 14;51(32):6312-9. Epub 2012 Aug 3. PMID:22849347 doi:http://dx.doi.org/10.1021/bi300849c
- ↑ 4.0 4.1 Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998 Jul 3;281(5373):108-11. PMID:9651244
- ↑ 5.0 5.1 Gittenberger-de Groot AC, Bartelings MM, Deruiter MC, Poelmann RE. Basics of cardiac development for the understanding of congenital heart malformations. Pediatr Res. 2005 Feb;57(2):169-76. doi: 10.1203/01.PDR.0000148710.69159.61. Epub, 2004 Dec 20. PMID:15611355 doi:http://dx.doi.org/10.1203/01.PDR.0000148710.69159.61
- ↑ Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006 Sep 29;313(5795):1922-7. doi: 10.1126/science.1132292. PMID:17008524 doi:http://dx.doi.org/10.1126/science.1132292
- ↑ Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacol Ther. 2005 Aug;107(2):252-68. doi: 10.1016/j.pharmthera.2005.03.005. PMID:15925411 doi:http://dx.doi.org/10.1016/j.pharmthera.2005.03.005
- ↑ Toko H, Zhu W, Takimoto E, Shiojima I, Hiroi Y, Zou Y, Oka T, Akazawa H, Mizukami M, Sakamoto M, Terasaki F, Kitaura Y, Takano H, Nagai T, Nagai R, Komuro I. Csx/Nkx2-5 is required for homeostasis and survival of cardiac myocytes in the adult heart. J Biol Chem. 2002 Jul 5;277(27):24735-43. doi: 10.1074/jbc.M107669200. Epub 2002 , Mar 11. PMID:11889119 doi:http://dx.doi.org/10.1074/jbc.M107669200
- ↑ McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003 Nov 5;42(9):1650-5. doi: 10.1016/j.jacc.2003.05.004. PMID:14607454 doi:http://dx.doi.org/10.1016/j.jacc.2003.05.004
- ↑ Carlson, B. M. (1994). Human embryology and developmental biology. St. Louis: Mosby.
- ↑ Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999 Mar;126(6):1269-80. doi: 10.1242/dev.126.6.1269. PMID:10021345 doi:http://dx.doi.org/10.1242/dev.126.6.1269
- ↑ Furtado MB, Wilmanns JC, Chandran A, Tonta M, Biben C, Eichenlaub M, Coleman HA, Berger S, Bouveret R, Singh R, Harvey RP, Ramialison M, Pearson JT, Parkington HC, Rosenthal NA, Costa MW. A novel conditional mouse model for Nkx2-5 reveals transcriptional regulation of cardiac ion channels. Differentiation. 2016 Jan-Mar;91(1-3):29-41. doi: 10.1016/j.diff.2015.12.003., Epub 2016 Feb 17. PMID:26897459 doi:http://dx.doi.org/10.1016/j.diff.2015.12.003
