Journal:JBIC:24
From Proteopedia
(Difference between revisions)

| (17 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene='' caption=''> | + | <StructureSection load='2m5u_pre.pdb' size='450' side='right' scene='57/573995/Cv/1' caption=''> |
=== Solution structure and metal ion binding sites of the human CPEB3 ribozyme's P4 domain === | === Solution structure and metal ion binding sites of the human CPEB3 ribozyme's P4 domain === | ||
| - | <big>Roland Sigel</big> <ref> | + | <big>Miriam Skilandat, Magdalena Rowinska-Zyrek and Roland K. O. Sigel</big> <ref>PMID 24652468 </ref> |
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
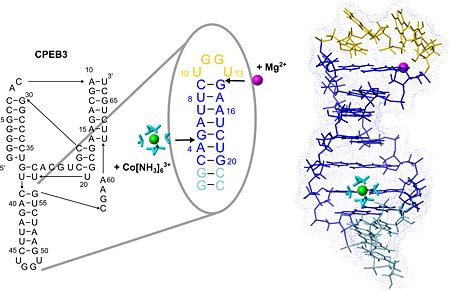
| + | The structure of the P4 hairpin provides the first structural information on the solution structure of the CPEB3 ribozyme, a highly conserved self-cleaving RNA sequence encoded in the genome of humans and other mammals. CPEB3 is one of very few examples of small ribozymes occuring in human and the only ribozyme that is suggested to be of human origin. Its function, however, is not yet understood. | ||
| + | The P4 hairpin points out of the complex interlaced structure of the CPEB3 ribozyme, it consists of a helix adopting the regular fold typical for RNA duplexes and is capped by a UGGU tetraloop. | ||
| + | [[Image:JBIC241.jpg|left|450px|thumb|The secondary structure of the CPEB3 ribozyme and the binding sites of Mg2+ and hexammine(III) cobalt that is an NMR-detectable mimic of a hexahydrated Mg2+ ion. P4 hairpin structure is shown with the two suggested metal-ion binding sites.]] | ||
| + | {{Clear}} | ||
| + | <scene name='57/573995/Fig/8'>P4 contains two well-defined binding sites for Magnesium(II)</scene> (see also image above) ions giving an excellent example of the two different binding modes of Mg(II) to RNA, which are direct coordination and coordination via a shell of water molecules. One of these binding sites probably confers extra stability to the fold of the tetraloop while the other one may play a role in stabilizing the adjacent active site region of the CPEB3 ribozyme. | ||
| + | The structure of the loop nucleotides is <scene name='57/573995/Fig/9'>exceptionally well defined as indicated by the very good convergence</scene> in this region. <scene name='57/573995/Fig/4'>Stacking interactions</scene> within this new tetraloop structure and a <scene name='57/573995/Fig/10'>putative hydrogen bond</scene> stabilize the hairpin fold which is similar to the AGUU tetraloop structure serving as a protein-binding motif in another RNA. Due to its exposed position and stable fold P4 could serve as an interaction site of the CPEB3 ribozyme for other biomolecules involved in regulating ribozymatic activity. | ||
| + | |||
| + | '''PDB reference:''' NMR structure of the P4 hairpin of the CPEB3 ribozyme, [[2m5u]]. | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
- ↑ Skilandat M, Rowinska-Zyrek M, Sigel RK. Solution structure and metal ion binding sites of the human CPEB3 ribozyme's P4 domain. J Biol Inorg Chem. 2014 Mar 21. PMID:24652468 doi:http://dx.doi.org/10.1007/s00775-014-1125-6
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.