Journal:JBIC:24
From Proteopedia
(Difference between revisions)

| (One intermediate revision not shown.) | |||
| Line 12: | Line 12: | ||
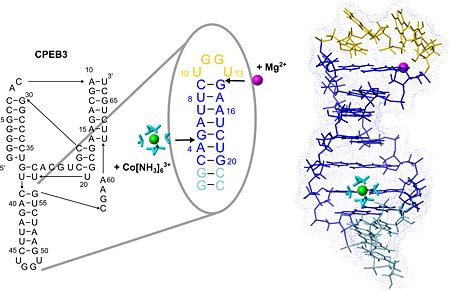
The structure of the loop nucleotides is <scene name='57/573995/Fig/9'>exceptionally well defined as indicated by the very good convergence</scene> in this region. <scene name='57/573995/Fig/4'>Stacking interactions</scene> within this new tetraloop structure and a <scene name='57/573995/Fig/10'>putative hydrogen bond</scene> stabilize the hairpin fold which is similar to the AGUU tetraloop structure serving as a protein-binding motif in another RNA. Due to its exposed position and stable fold P4 could serve as an interaction site of the CPEB3 ribozyme for other biomolecules involved in regulating ribozymatic activity. | The structure of the loop nucleotides is <scene name='57/573995/Fig/9'>exceptionally well defined as indicated by the very good convergence</scene> in this region. <scene name='57/573995/Fig/4'>Stacking interactions</scene> within this new tetraloop structure and a <scene name='57/573995/Fig/10'>putative hydrogen bond</scene> stabilize the hairpin fold which is similar to the AGUU tetraloop structure serving as a protein-binding motif in another RNA. Due to its exposed position and stable fold P4 could serve as an interaction site of the CPEB3 ribozyme for other biomolecules involved in regulating ribozymatic activity. | ||
| + | '''PDB reference:''' NMR structure of the P4 hairpin of the CPEB3 ribozyme, [[2m5u]]. | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
- ↑ Skilandat M, Rowinska-Zyrek M, Sigel RK. Solution structure and metal ion binding sites of the human CPEB3 ribozyme's P4 domain. J Biol Inorg Chem. 2014 Mar 21. PMID:24652468 doi:http://dx.doi.org/10.1007/s00775-014-1125-6
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.