Triose Phosphate Isomerase Structure & Mechanism
From Proteopedia
(Difference between revisions)
| (21 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | <StructureSection load='1hti' size='300' side='right' scene='' caption='Human triosephosphate isomerase complex with phosphoglycolic acid [[1hti]]'> |
| + | |||
| + | [[Image:TriosePhosphateIsomerase_Ribbon_pastel_photo_small.jpg|thumb|left|300px|Ribbon drawing for one chain of the "TIM barrel" fold]] | ||
| + | <br /> | ||
===General Information=== | ===General Information=== | ||
| Line 5: | Line 8: | ||
===Structural Characteristics=== | ===Structural Characteristics=== | ||
| - | The secondary structure consists of 14 alpha helices and 8 beta sheets per monomer, making it fall in the SCOP category of alpha and beta proteins. The tertiary structure is a <scene name='Christian_Krenk_Sandbox/Alpha_beta_barrel/2'>alpha-beta barrel | + | The secondary structure consists of 14 alpha helices and 8 beta sheets per monomer, making it fall in the SCOP category of alpha and beta proteins. The tertiary structure is a <scene name='Christian_Krenk_Sandbox/Alpha_beta_barrel/2'>alpha-beta barrel</scene>, and it is the prototypical example of the "TIM barrel" fold (see ribbon drawing). |
The quaternary structure is a homodimer. The molecular weight of the enzyme is estimated at 57,400 Da.<ref name= "dab">PMID:752201</ref> | The quaternary structure is a homodimer. The molecular weight of the enzyme is estimated at 57,400 Da.<ref name= "dab">PMID:752201</ref> | ||
| - | |||
| - | |||
| - | |||
| - | |||
===Mechanism=== | ===Mechanism=== | ||
| Line 17: | Line 16: | ||
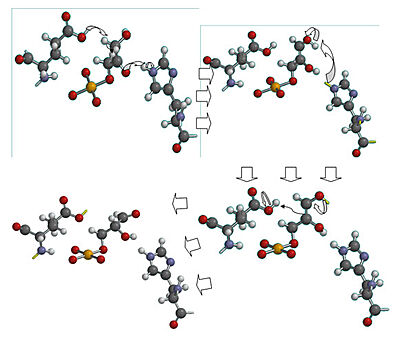
<scene name='Christian_Krenk_Sandbox/Active_site/1'> Glu 165 acts as the base and grabs the C2 proton on glyceraldehyde-3-phosphate, while His 95 is H-bonded to the carbonyl oxygen and acts as the acid by protonating carbonyl oxygen.</scene> The enediol intermediate is negatively charged, but is somewhat <scene name='Christian_Krenk_Sandbox/Lysine/1'>stabilized by the positively charged side chain of Lys 12.</scene> <ref name= "lodi">PMID:8130193</ref> Mutation of Lys 12 to Arg increases Km by a factor of 22 and decreases Vmax by a factor of 180.<ref name="lodi" /> To convert the enediol intermediate to DHAP, C1 is protonated by Glu 165, with His 95 removing a proton from C2’s OH group. As a result, the catalytic groups are back to their original states, and catalysis is complete. With GAP as a substrate, Km for the reaction is .34 mM and Vmax is 7200 units/mg protein at 25 degrees C and pH 7.5.<ref name= "dab" /> | <scene name='Christian_Krenk_Sandbox/Active_site/1'> Glu 165 acts as the base and grabs the C2 proton on glyceraldehyde-3-phosphate, while His 95 is H-bonded to the carbonyl oxygen and acts as the acid by protonating carbonyl oxygen.</scene> The enediol intermediate is negatively charged, but is somewhat <scene name='Christian_Krenk_Sandbox/Lysine/1'>stabilized by the positively charged side chain of Lys 12.</scene> <ref name= "lodi">PMID:8130193</ref> Mutation of Lys 12 to Arg increases Km by a factor of 22 and decreases Vmax by a factor of 180.<ref name="lodi" /> To convert the enediol intermediate to DHAP, C1 is protonated by Glu 165, with His 95 removing a proton from C2’s OH group. As a result, the catalytic groups are back to their original states, and catalysis is complete. With GAP as a substrate, Km for the reaction is .34 mM and Vmax is 7200 units/mg protein at 25 degrees C and pH 7.5.<ref name= "dab" /> | ||
| - | [[Image:ckrenkmechanism.jpg|left|thumb| | + | [[Image:ckrenkmechanism.jpg|left|thumb|400px| '''Mechanism of Triose phosphate isomerase'''. Created by Christian Krenk using Spartan 08.]] |
An interesting part of the enzyme is the <scene name='Christian_Krenk_Sandbox/Flexible_loop/1'>flexible loop</scene> that stabilizes the enediol-like transition state. The flexible loop (residues 167-176)<ref>PMID:2204418</ref> closes over the active site like a hinged lid when substrate is bound, thus preventing phosphate from leaving. A four-residue segment of the loop H-bonds with the phosphate group of the substrate.<ref name="book" /> Without the loop, the enediol intermediate would eliminate phosphate, with the end products being inorganic phosphate and toxic methylglyoxal.<ref name="book" /> | An interesting part of the enzyme is the <scene name='Christian_Krenk_Sandbox/Flexible_loop/1'>flexible loop</scene> that stabilizes the enediol-like transition state. The flexible loop (residues 167-176)<ref>PMID:2204418</ref> closes over the active site like a hinged lid when substrate is bound, thus preventing phosphate from leaving. A four-residue segment of the loop H-bonds with the phosphate group of the substrate.<ref name="book" /> Without the loop, the enediol intermediate would eliminate phosphate, with the end products being inorganic phosphate and toxic methylglyoxal.<ref name="book" /> | ||
| + | ==3D structures of triose phosphate isomerase== | ||
| - | + | [[Triose Phosphate Isomerase]] | |
| - | + | ||
| - | + | ||
| + | ==Additional Resources== | ||
| + | For additional information, see: [[Carbohydrate Metabolism]] | ||
| + | <br /> | ||
| + | </StructureSection> | ||
===References=== | ===References=== | ||
<references/> | <references/> | ||
| + | 7. Wierenga RK, Kapetaniou EG, Venkatesan R. Triosephosphate isomerase: a highly evolved biocatalyst. Cellular and Molecular Life Sciences. 2010 August 7 67:3961-3982. PMID: 20694739 <ref>PMID:20694739</ref> | ||
Current revision
| |||||||||||
References
- ↑ Kinoshita T, Maruki R, Warizaya M, Nakajima H, Nishimura S. Structure of a high-resolution crystal form of human triosephosphate isomerase: improvement of crystals using the gel-tube method. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 Apr 1;61(Pt, 4):346-9. Epub 2005 Mar 24. PMID:16511037 doi:10.1107/S1744309105008341
- ↑ Mande SC, Mainfroid V, Kalk KH, Goraj K, Martial JA, Hol WG. Crystal structure of recombinant human triosephosphate isomerase at 2.8 A resolution. Triosephosphate isomerase-related human genetic disorders and comparison with the trypanosomal enzyme. Protein Sci. 1994 May;3(5):810-21. PMID:8061610
- ↑ 3.0 3.1 Dabrowska A, Kamrowska I, Baranowski T. Purification, crystallization and properties of triosephosphate isomerase from human skeletal muscle. Acta Biochim Pol. 1978;25(3):247-56. PMID:752201
- ↑ 4.0 4.1 4.2 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. p. 495. Print.
- ↑ 5.0 5.1 Lodi PJ, Chang LC, Knowles JR, Komives EA. Triosephosphate isomerase requires a positively charged active site: the role of lysine-12. Biochemistry. 1994 Mar 15;33(10):2809-14. PMID:8130193
- ↑ Lolis E, Petsko GA. Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-A resolution: implications for catalysis. Biochemistry. 1990 Jul 17;29(28):6619-25. PMID:2204418
7. Wierenga RK, Kapetaniou EG, Venkatesan R. Triosephosphate isomerase: a highly evolved biocatalyst. Cellular and Molecular Life Sciences. 2010 August 7 67:3961-3982. PMID: 20694739 [1]
Proteopedia Page Contributors and Editors (what is this?)
Christian Krenk, Alexander Berchansky, Diamond B. Reese, Michal Harel, Jane S. Richardson, David Canner