We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Pyruvate Kinase
From Proteopedia
(Difference between revisions)
| (16 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | |||
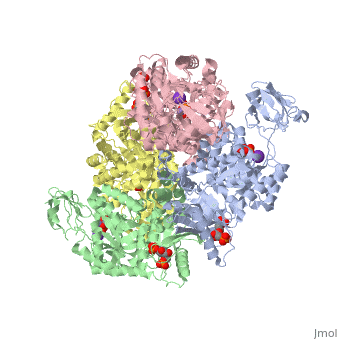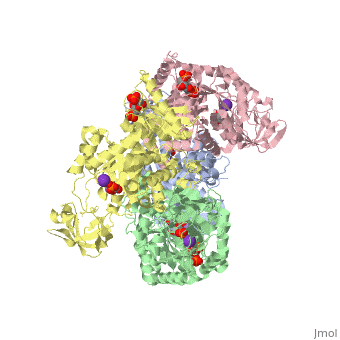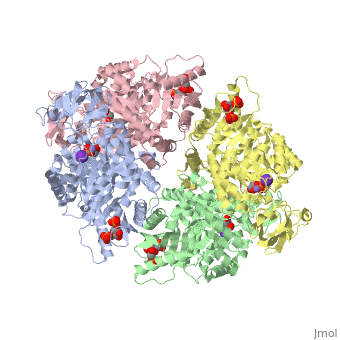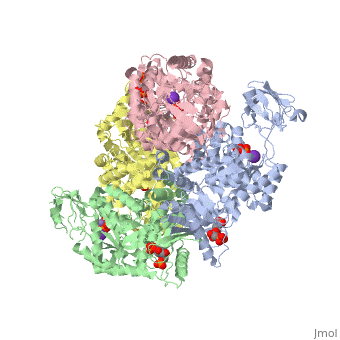
<StructureSection load='2vgb' size='350' side='right' scene='' caption='Human pyruvate kinase tetramer complex with fructose diphosphate, phosphoglycolic acid, Mn+2 and K+ (purple) ions (PDB code [[2vgb]])'> | <StructureSection load='2vgb' size='350' side='right' scene='' caption='Human pyruvate kinase tetramer complex with fructose diphosphate, phosphoglycolic acid, Mn+2 and K+ (purple) ions (PDB code [[2vgb]])'> | ||
[[Pyruvate Kinase]] is an enzyme that is involved in glycolysis. Pyruvate kinase’s function is to catalyze the last step of glycolysis; thereby, generating the second ATP of glycolysis and pyruvate. It is able to catalyze this step by transferring the phosphate group from phosphoenolpyruvate (PEP) to ADP <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=501|}}</ref>. See [[Glycolysis Enzymes]]. | [[Pyruvate Kinase]] is an enzyme that is involved in glycolysis. Pyruvate kinase’s function is to catalyze the last step of glycolysis; thereby, generating the second ATP of glycolysis and pyruvate. It is able to catalyze this step by transferring the phosphate group from phosphoenolpyruvate (PEP) to ADP <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=501|}}</ref>. See [[Glycolysis Enzymes]]. | ||
| Line 6: | Line 5: | ||
This particular protein is found in Homo sapiens and has the abbreviation PK. Pyruvate kinase belongs to the all beta proteins class and has the PK beta-barrel domain-like fold. It belongs to the PK beta-barrel domain-like superfamily and pyruvate kinase beta-barrel domain family<ref>{{website| title=SCOP: Protein: Pyruvate kinase (PK) from Human (Homo sapiens) [TaxId: 9606]|url=http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html|}}</ref>. | This particular protein is found in Homo sapiens and has the abbreviation PK. Pyruvate kinase belongs to the all beta proteins class and has the PK beta-barrel domain-like fold. It belongs to the PK beta-barrel domain-like superfamily and pyruvate kinase beta-barrel domain family<ref>{{website| title=SCOP: Protein: Pyruvate kinase (PK) from Human (Homo sapiens) [TaxId: 9606]|url=http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html|}}</ref>. | ||
| - | Though pyruvate kinase is classified into all beta proteins, pyruvate kinase's <scene name='Keegan_Gelvoria_Sandbox_1/Secondary_structure/1'>secondary structure</scene> comprises of both alpha helices and beta sheets. In the quaternary structure of pyruvate kinase, it can be observed to have <scene name='Keegan_Gelvoria_Sandbox_1/Structure_4_domains/1'>four domains</scene> in humans. Thus, this enzyme is tetrameric with <scene name='Keegan_Gelvoria_Sandbox_1/Metal_binding_sites/1'>metal binding sites</scene> on each domain for the <scene name='Keegan_Gelvoria_Sandbox_1/Ligands/1'>K+</scene> and Mg2+ ligands to bind to. There are four types of tissue-specific isozymes: L (liver), R (red blood cells), M1 (muscle, heart, and brain), and M2 (early fetal tissue)<ref>{{website| title=SCOP: Protein: Pyruvate kinase (PK) from Human (Homo sapiens) [TaxId: 9606]|url=http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html|}}</ref>. | + | Though pyruvate kinase is classified into all beta proteins, pyruvate kinase's <scene name='Keegan_Gelvoria_Sandbox_1/Secondary_structure/1'>secondary structure</scene> comprises of both alpha helices and beta sheets. In the quaternary structure of pyruvate kinase, it can be observed to have <scene name='Keegan_Gelvoria_Sandbox_1/Structure_4_domains/1'>four domains</scene> in humans. Thus, this enzyme is tetrameric with <scene name='Keegan_Gelvoria_Sandbox_1/Metal_binding_sites/1'>metal binding sites</scene> on each domain for the <scene name='Keegan_Gelvoria_Sandbox_1/Ligands/1'>K+</scene> and Mg2+ ligands to bind to. There are four types of tissue-specific isozymes: '''L''' (liver), '''R''' (red blood cells), '''M1''' (muscle, heart, and brain), and '''M2''' (early fetal tissue)<ref>{{website| title=SCOP: Protein: Pyruvate kinase (PK) from Human (Homo sapiens) [TaxId: 9606]|url=http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html|}}</ref>. |
==Mechanism== | ==Mechanism== | ||
| - | + | ||
| - | + | ||
<scene name='Keegan_Gelvoria_Sandbox_1/N_c_rainbow/null'>Pyruvate Kinase</scene> catalyzes the final reaction of glycolysis. It couples the free energy of PEP cleavage to the generation of ATP during the synthesis of the final product, pyruvate. This reaction necessitates one K+ and two Mg2+ cations to be used in two steps. The first step is the nucleophilic attack of the PEP phosphorous atom by β-phosphoryl oxygen of ADP; this step displaces enolpyruvate while forming ATP. In the second step, enolpyruvate tautomerizes to pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=502|}}</ref>. The formation of a high-energy intermediate by enolase in the 9th reaction of glycolysis allows for the synthesis of ATP in this reaction. Though the hydrolysis of 2PG is insufficient in driving the synthesis of ATP, the dehydration of 2PG allows for such a reaction to occur by forming a high-energy intermediate. The high potential of PEP reflects the large release of energy that occurs with the conversion of enolpyruvate to its keto tautomer, pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=503|}}</ref>. | <scene name='Keegan_Gelvoria_Sandbox_1/N_c_rainbow/null'>Pyruvate Kinase</scene> catalyzes the final reaction of glycolysis. It couples the free energy of PEP cleavage to the generation of ATP during the synthesis of the final product, pyruvate. This reaction necessitates one K+ and two Mg2+ cations to be used in two steps. The first step is the nucleophilic attack of the PEP phosphorous atom by β-phosphoryl oxygen of ADP; this step displaces enolpyruvate while forming ATP. In the second step, enolpyruvate tautomerizes to pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=502|}}</ref>. The formation of a high-energy intermediate by enolase in the 9th reaction of glycolysis allows for the synthesis of ATP in this reaction. Though the hydrolysis of 2PG is insufficient in driving the synthesis of ATP, the dehydration of 2PG allows for such a reaction to occur by forming a high-energy intermediate. The high potential of PEP reflects the large release of energy that occurs with the conversion of enolpyruvate to its keto tautomer, pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=503|}}</ref>. | ||
| Line 42: | Line 40: | ||
Red blood cells, in a state of pyruvate kinase deficiency, rapidly become deficient in ATP and can undergo hemolysis.This is transmitted as an autosomal recessive trit. The severity of hemolysis is extremely variable such as a mild case to life-threatening neonatal anaemia requiring transfusions. Over one hundred eighty different mutations have been discovered in relation to this deficiency with most being autosomal recessive, but a few strands are autosomal dominant. The deficiency causes red blood cells to deform into echinocytes on peripheral blood smears. This causes the buildup of reaction intermediates which can also increase the level of 2,3-bisphosphoglycerate in the cells. This causes a rightward shift in the hemoglobin oxygen saturation curve, which means that there is a decreased oxygen affinity for the hemoglobin and earlier oxygen unloading than under normal conditions | Red blood cells, in a state of pyruvate kinase deficiency, rapidly become deficient in ATP and can undergo hemolysis.This is transmitted as an autosomal recessive trit. The severity of hemolysis is extremely variable such as a mild case to life-threatening neonatal anaemia requiring transfusions. Over one hundred eighty different mutations have been discovered in relation to this deficiency with most being autosomal recessive, but a few strands are autosomal dominant. The deficiency causes red blood cells to deform into echinocytes on peripheral blood smears. This causes the buildup of reaction intermediates which can also increase the level of 2,3-bisphosphoglycerate in the cells. This causes a rightward shift in the hemoglobin oxygen saturation curve, which means that there is a decreased oxygen affinity for the hemoglobin and earlier oxygen unloading than under normal conditions | ||
<ref>PMID:17360088</ref>. | <ref>PMID:17360088</ref>. | ||
| - | + | ||
==3D structures of pyruvate kinase== | ==3D structures of pyruvate kinase== | ||
| + | [[Pyruvate kinase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Additional Resources== | ==Additional Resources== | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ authors, The scop. "Structural Classification of Proteins". 2009. 2/26 2010. <http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html>.
- ↑ authors, The scop. "Structural Classification of Proteins". 2009. 2/26 2010. <http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html>.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ Dann LG, Britton HG. Kinetics and mechanism of action of muscle pyruvate kinase. Biochem J. 1978 Jan 1;169(1):39-54. PMID:629752
- ↑ Mattevi A, Bolognesi M, Valentini G. The allosteric regulation of pyruvate kinase. FEBS Lett. 1996 Jun 24;389(1):15-9. PMID:8682196
- ↑ Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998 Feb 15;6(2):195-210. PMID:9519410
- ↑ Oria-Hernandez J, Cabrera N, Perez-Montfort R, Ramirez-Silva L. Pyruvate kinase revisited: the activating effect of K+. J Biol Chem. 2005 Nov 11;280(45):37924-9. Epub 2005 Sep 7. PMID:16147999 doi:10.1074/jbc.M508490200
- ↑ Dann LG, Britton HG. Kinetics and mechanism of action of muscle pyruvate kinase. Biochem J. 1978 Jan 1;169(1):39-54. PMID:629752
- ↑ Oria-Hernandez J, Cabrera N, Perez-Montfort R, Ramirez-Silva L. Pyruvate kinase revisited: the activating effect of K+. J Biol Chem. 2005 Nov 11;280(45):37924-9. Epub 2005 Sep 7. PMID:16147999 doi:10.1074/jbc.M508490200
- ↑ Zanella A, Fermo E, Bianchi P, Chiarelli LR, Valentini G. Pyruvate kinase deficiency: the genotype-phenotype association. Blood Rev. 2007 Jul;21(4):217-31. Epub 2007 Mar 13. PMID:17360088 doi:10.1016/j.blre.2007.01.001
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Keegan Gelvoria, David Canner, Ann Taylor, Andrew Alexander