Pyruvate Kinase
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 8: | Line 8: | ||
==Mechanism== | ==Mechanism== | ||
| - | + | ||
| - | + | ||
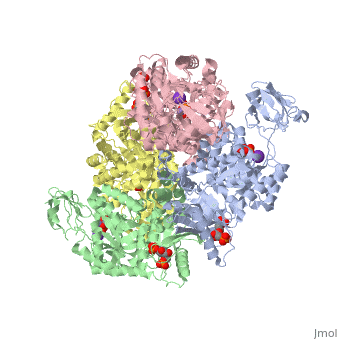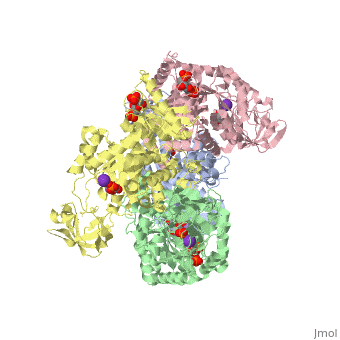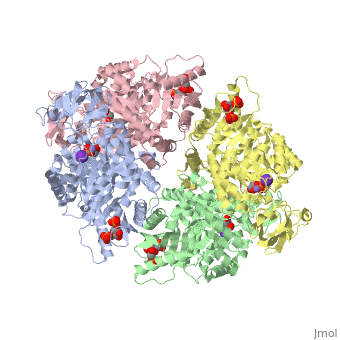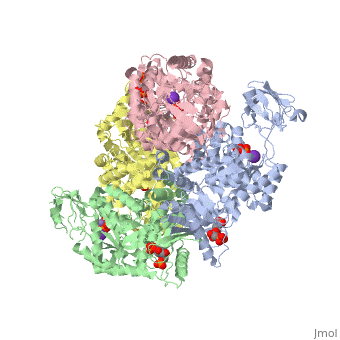
<scene name='Keegan_Gelvoria_Sandbox_1/N_c_rainbow/null'>Pyruvate Kinase</scene> catalyzes the final reaction of glycolysis. It couples the free energy of PEP cleavage to the generation of ATP during the synthesis of the final product, pyruvate. This reaction necessitates one K+ and two Mg2+ cations to be used in two steps. The first step is the nucleophilic attack of the PEP phosphorous atom by β-phosphoryl oxygen of ADP; this step displaces enolpyruvate while forming ATP. In the second step, enolpyruvate tautomerizes to pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=502|}}</ref>. The formation of a high-energy intermediate by enolase in the 9th reaction of glycolysis allows for the synthesis of ATP in this reaction. Though the hydrolysis of 2PG is insufficient in driving the synthesis of ATP, the dehydration of 2PG allows for such a reaction to occur by forming a high-energy intermediate. The high potential of PEP reflects the large release of energy that occurs with the conversion of enolpyruvate to its keto tautomer, pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=503|}}</ref>. | <scene name='Keegan_Gelvoria_Sandbox_1/N_c_rainbow/null'>Pyruvate Kinase</scene> catalyzes the final reaction of glycolysis. It couples the free energy of PEP cleavage to the generation of ATP during the synthesis of the final product, pyruvate. This reaction necessitates one K+ and two Mg2+ cations to be used in two steps. The first step is the nucleophilic attack of the PEP phosphorous atom by β-phosphoryl oxygen of ADP; this step displaces enolpyruvate while forming ATP. In the second step, enolpyruvate tautomerizes to pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=502|}}</ref>. The formation of a high-energy intermediate by enolase in the 9th reaction of glycolysis allows for the synthesis of ATP in this reaction. Though the hydrolysis of 2PG is insufficient in driving the synthesis of ATP, the dehydration of 2PG allows for such a reaction to occur by forming a high-energy intermediate. The high potential of PEP reflects the large release of energy that occurs with the conversion of enolpyruvate to its keto tautomer, pyruvate <ref>{{book |author=Voet, Donald; Voet, Judith C.; Pratt, Charlotte W.|title=Fundamentals of Biochemistry: Life at the Molecular Level|edition= 3|pages=503|}}</ref>. | ||
| Line 46: | Line 45: | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of pyruvate kinase== | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *Pyruvate kinase | ||
| - | |||
| - | **[[3bjf]], [[3bjt]], [[1t5a]], [[1zjh]], [[3srf]], [[3srh]] - hPyK M1/M2 - human<br /> | ||
| - | **[[4wj8]], [[4qg6]], [[4qg8]], [[4qg9]], [[4qgc]], [[3g2g]], [[4yj5]], [[6b6u]] - hPyK M1/M2 (mutant)<br /> | ||
| - | **[[2vgb]] – hPyK R/L<br /> | ||
| - | **[[2vgf]], [[2vgg]], [[2vgi]] - hPyK R/L (mutant) <br /> | ||
| - | **[[6nn5]], [[6nn7]], [[6nn8]] – hPyK PKLR (mutant)<br /> | ||
| - | **[[1pkm]], [[1pyk]] – PyK – cat<br /> | ||
| - | **[[1f3w]] – rPyK – rabbit<br /> | ||
| - | **[[1f3x]] - rPyK (mutant) <br /> | ||
| - | **[[1pky]], [[4yng]] – EcPyK - ''Eschericia coli''<br /> | ||
| - | **[[1e0t]], [[1e0u]] - EcPyK (mutant) <br /> | ||
| - | **[[3t05]], [[3t0t]] – SaPyK – ''Staphylococcus aureus''<br /> | ||
| - | **[[3qtg]] – PyK – ''Pyrobaculum aerophilum''<br /> | ||
| - | **[[4krz]] - TcPyK – ''Trypanosoma cruzei''<br /> | ||
| - | **[[5wrp]] - MtPyK – ''Mycobacterium tuberculosis''<br /> | ||
| - | **[[3hqn]], [[3e0w]], [[1pkl]] – LmPyK – ''Leishmania mexicana''<br /> | ||
| - | **[[3khd]] – PyK – ''Plasmodium falciparum''<br /> | ||
| - | **[[3gg8]], [[3eoe]] – PyK – ''Toxoplasma gondii''<br /> | ||
| - | **[[3ma8]], [[4drs]] – PyK – ''Cryptosporidium parvum''<br /> | ||
| - | **[[2e28]] - PyK (mutant) – ''Geobacillus stearothermophilus''<br /> | ||
| - | |||
| - | *Pyruvate kinase binary complex | ||
| - | |||
| - | **[[4b2d]] - hPyK M1/M2 + fructose bisphosphate<br /> | ||
| - | **[[4rpp]] - hPyK M2 (mutant) + fructose bisphosphate<br /> | ||
| - | **[[4fxj]] - hPyK M1/M2 + phenylalanine<br /> | ||
| - | **[[6gg3]] - hPyK M2 + alanine<br /> | ||
| - | **[[6gg4]] - hPyK M2 + phenylalanine<br /> | ||
| - | **[[6gg5]] - hPyK M2 + tryptophan<br /> | ||
| - | **[[6gg6]] - hPyK M2 + serine<br /> | ||
| - | **[[6nub]] - hPyK M2 (mutant) + serine<br /> | ||
| - | **[[6nu1]] - hPyK M2 + cysteine<br /> | ||
| - | **[[6nu5]] - hPyK M2 (mutant) + cysteine<br /> | ||
| - | **[[6ech]], [[6eck]] - PyK PKLR + fructose bisphosphate - rat<br /> | ||
| - | **[[5x1v]], [[5x1w]] - hPyK M2 + inhibitor<br /> | ||
| - | **[[4rpp]] - hPyK M2 (mutant) + fructose bisphosphate<br />**[[3qv9]] – TcPyK + Ponceau S <br /> | ||
| - | **[[3qv6]] – LmPyK + acid blue 80<br /> | ||
| - | **[[3e0v]] – LmPyK + sulfate<br /> | ||
| - | **[[3hqq]] - LmPyK + fructose bisphosphate<br /> | ||
| - | **[[3qv8]] – LmPyK + benzothiazole<br /> | ||
| - | **[[3pp7]] – LmPyK + suramin<br /> | ||
| - | **[[3is4]], [[3ktx]] – LmPyK + pyrenetetrasulfonic acid<br /> | ||
| - | **[[3t07]] – SaPyK + bis-indole alkaloid<br /> | ||
| - | **[[4hyw]] – TbPyK + fructose bisphosphate – ''Trypanosoma brucei''<br /> | ||
| - | **[[5ws8]] - MtPyK + oxalate<br /> | ||
| - | **[[6p0y]] - CpPyK + ADP<br /> | ||
| - | |||
| - | *Pyruvate kinase higher complex | ||
| - | |||
| - | **[[3me3]] – hPyK M1/M2 + aniline derivative + fructose bisphosphate<br /> | ||
| - | **[[4g1n]] - hPyK M1/M2 + pyridazine derivative + oxalate<br /> | ||
| - | **[[3h6o]] - hPyK M1/M2 + pyridazine derivative + fructose bisphosphate<br /> | ||
| - | **[[3gqy]], [[3gr4]], [[3u2z]] - hPyK M1/M2 + piperazine derivative + fructose bisphosphate<br /> | ||
| - | **[[5x0i]] - hPyK M2 (mutant) + serine + fructose bisphosphate<br /> | ||
| - | **[[5x1v]], [[5x1w]] - hPyK M2 + activator + fructose bisphosphate<br /> | ||
| - | **[[4jpg]] - hPyK M1/M2 + pyrimidine derivative + fructose bisphosphate<br /> | ||
| - | **[[3srd]] - hPyK M1/M2 + oxalate + fructose bisphosphate<br /> | ||
| - | **[[4fxf]] - hPyK M1/M2 + oxalate + ATP + fructose bisphosphate<br /> | ||
| - | **[[4ima]], [[4ip7]] - hPyK L (mutant) + citrate + adenosine + fructose bisphosphate<br /> | ||
| - | **[[3n25]] – rPyK M1/M2 + proline + Mn + pyruvate<br /> | ||
| - | **[[2g50]] - rPyK M1/M2 + alanine + Mn + pyruvate<br /> | ||
| - | **[[1pkn]] - rPyK + Mn + pyruvate<br /> | ||
| - | **[[1aqf]] – rPyK + Mg + phospholactate<br /> | ||
| - | **[[1a49]], [[1a5u]] - rPyK + ATP + oxalate<br /> | ||
| - | **[[1a3w]] – yPyK + Mn + phosphoglycolic acid + fructose bisphosphate – yeast<br /> | ||
| - | **[[1a3x]] - yPyK + Mn + phosphoglycolic acid<br /> | ||
| - | **[[4hyv]] – TbPyK + phosphoenolpyruvate + fructose bisphosphate <br /> | ||
| - | **[[4kct]] – TbPyK + pyruvate + fructose bisphosphate <br /> | ||
| - | **[[4kcu]] – TbPyK + malate + fructose bisphosphate <br /> | ||
| - | **[[4kcv]] – TbPyK + oxoglutarate + fructose bisphosphate <br /> | ||
| - | **[[4kcw]] – TbPyK + oxalate + fructose bisphosphate <br /> | ||
| - | **[[4ks0]] – TcPyK + oxalate + fructose bisphosphate <br /> | ||
| - | **[[5ws9]] – MtPyK + oxalate + AMP + ATP <br /> | ||
| - | **[[5wsa]] – MtPyK + oxalate + glucose-6-phosphate <br /> | ||
| - | **[[5wsb]], [[5wsc]] – MtPyK + oxalate + glucose-6-phosphate + AMP<br /> | ||
| - | **[[6ito]] – MtPyK + oxalate + ribose-5-phosphate + AMP<br /> | ||
| - | **[[3hqo]] – LmPyK + ATP + oxalate<br /> | ||
| - | **[[3hqp]] - LmPyK + ATP + oxalate + fructose bisphosphate<br /> | ||
| - | **[[3qv7]] – LmPyK + acid blue 25 + Ponceau S<br /> | ||
| - | **[[3srk]] – LmPyK + saccharine + inhibitor<br /> | ||
| - | **[[6qxl]] – PyK + malonate + glucose-6-phosphate + Mg – ''Pseudomonas aerugnosa''<br /> | ||
| - | |||
| - | }} | ||
| - | |||
| - | |||
| - | |||
==Additional Resources== | ==Additional Resources== | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ authors, The scop. "Structural Classification of Proteins". 2009. 2/26 2010. <http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html>.
- ↑ authors, The scop. "Structural Classification of Proteins". 2009. 2/26 2010. <http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html>.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ Dann LG, Britton HG. Kinetics and mechanism of action of muscle pyruvate kinase. Biochem J. 1978 Jan 1;169(1):39-54. PMID:629752
- ↑ Mattevi A, Bolognesi M, Valentini G. The allosteric regulation of pyruvate kinase. FEBS Lett. 1996 Jun 24;389(1):15-9. PMID:8682196
- ↑ Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998 Feb 15;6(2):195-210. PMID:9519410
- ↑ Oria-Hernandez J, Cabrera N, Perez-Montfort R, Ramirez-Silva L. Pyruvate kinase revisited: the activating effect of K+. J Biol Chem. 2005 Nov 11;280(45):37924-9. Epub 2005 Sep 7. PMID:16147999 doi:10.1074/jbc.M508490200
- ↑ Dann LG, Britton HG. Kinetics and mechanism of action of muscle pyruvate kinase. Biochem J. 1978 Jan 1;169(1):39-54. PMID:629752
- ↑ Oria-Hernandez J, Cabrera N, Perez-Montfort R, Ramirez-Silva L. Pyruvate kinase revisited: the activating effect of K+. J Biol Chem. 2005 Nov 11;280(45):37924-9. Epub 2005 Sep 7. PMID:16147999 doi:10.1074/jbc.M508490200
- ↑ Zanella A, Fermo E, Bianchi P, Chiarelli LR, Valentini G. Pyruvate kinase deficiency: the genotype-phenotype association. Blood Rev. 2007 Jul;21(4):217-31. Epub 2007 Mar 13. PMID:17360088 doi:10.1016/j.blre.2007.01.001
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Keegan Gelvoria, David Canner, Ann Taylor, Andrew Alexander