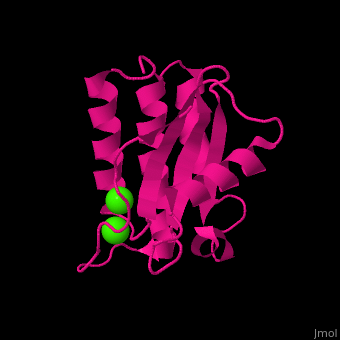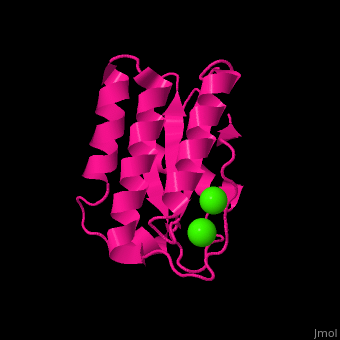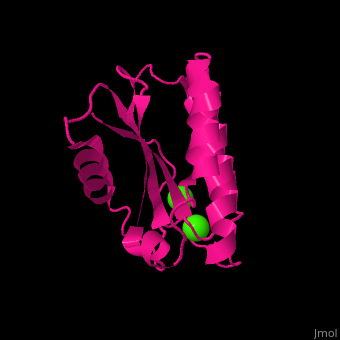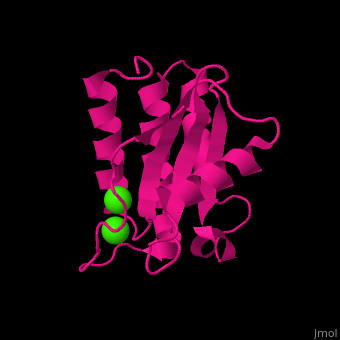We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
BA42
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==BA42 Protein from ''Bizionia argentinensis''== | ==BA42 Protein from ''Bizionia argentinensis''== | ||
| - | <StructureSection load=' | + | <StructureSection load='' size='400' side='right' caption='Antarctic bacterium protein BA42 complex with Ca2+ |
| + | ions (PDB code [[4oa3]])' scene='71/715464/Cv/1'> | ||
== Function == | == Function == | ||
| Line 7: | Line 8: | ||
Protein phosphatase from ''Bizionia argentinensis'' | Protein phosphatase from ''Bizionia argentinensis'' | ||
== Structural highlights == | == Structural highlights == | ||
| - | '''BA42''' belongs to the TPM protein family from Pfam. The TPM domain family is named after the three founding proteins TLP18.3, Psb32 and MOLO-1. TPM domains have a characteristic fold <scene name='71/715464/Cv/ | + | '''BA42''' belongs to the TPM protein family from Pfam. The TPM domain family is named after the three founding proteins TLP18.3, Psb32 and MOLO-1. TPM domains have a characteristic fold <scene name='71/715464/Cv/9'>(αβαβαββαα or βαβαββαα)</scene> composed of α helices (3+3<ref>pmid 21908686</ref> or 2+3<ref>pmid 22198206</ref>) flanking four central β strands. The TPM fold has not been found in other protein domains to date. TPM was previously referred to as "DUF477" and "Repair_PSII". |
In plants, the TPM domain-containing proteins TLP18.3 and Psb32 that have been implicated in the photosystem II (PSII) repair cycle. It may be involved in the regulation of synthesis/degradation of the D1 protein of the PSII core and in the assembly of PSII monomers into dimers in the grana stacks.<ref>pmid 17576201</ref> | In plants, the TPM domain-containing proteins TLP18.3 and Psb32 that have been implicated in the photosystem II (PSII) repair cycle. It may be involved in the regulation of synthesis/degradation of the D1 protein of the PSII core and in the assembly of PSII monomers into dimers in the grana stacks.<ref>pmid 17576201</ref> | ||
| Line 13: | Line 14: | ||
In the model nematode ''C. elegans'', the MOLO-1 protein is an auxiliary subunit that positively modulates the gating of levamisole-sensitive acetylcholine receptors.<ref>pmid 22922783</ref> | In the model nematode ''C. elegans'', the MOLO-1 protein is an auxiliary subunit that positively modulates the gating of levamisole-sensitive acetylcholine receptors.<ref>pmid 22922783</ref> | ||
| - | <scene name='71/715464/Cv/ | + | <scene name='71/715464/Cv/7'>1st Ca2+ coordination site</scene>. Water molecules sre shown as red spheres. |
| + | |||
| + | <scene name='71/715464/Cv/8'>2nd Ca2+ coordination site</scene> in Antarctic bacterium protein BA42 (PDB code [[4oa3]]).<ref>PMID:25116514</ref> | ||
</StructureSection> | </StructureSection> | ||
== 3D structure of BA42 == | == 3D structure of BA42 == | ||
| - | [[2mpb]] - BaBA42 - ''Bizonia argentinesis'' - NMR<br /> | + | [[2lt2]], [[2mpb]] - BaBA42 - ''Bizonia argentinesis'' - NMR<br /> |
[[4oa3]] - BaBA42<br /> | [[4oa3]] - BaBA42<br /> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
BA42 Protein from Bizionia argentinensis
| |||||||||||
3D structure of BA42
2lt2, 2mpb - BaBA42 - Bizonia argentinesis - NMR
4oa3 - BaBA42
References
- ↑ Wu HY, Liu MS, Lin TP, Cheng YS. Structural and Functional Assays of AtTLP18.3 Identify Its Novel Acid Phosphatase Activity in Thylakoid Lumen. Plant Physiol. 2011 Sep 9. PMID:21908686 doi:10.1104/pp.111.184739
- ↑ Eletsky A, Acton TB, Xiao R, Everett JK, Montelione GT, Szyperski T. Solution NMR structures reveal a distinct architecture and provide first structures for protein domain family PF04536. J Struct Funct Genomics. 2012 Mar;13(1):9-14. doi: 10.1007/s10969-011-9122-2., Epub 2011 Dec 24. PMID:22198206 doi:http://dx.doi.org/10.1007/s10969-011-9122-2
- ↑ Sirpio S, Allahverdiyeva Y, Suorsa M, Paakkarinen V, Vainonen J, Battchikova N, Aro EM. TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem J. 2007 Sep 15;406(3):415-25. PMID:17576201 doi:http://dx.doi.org/10.1042/BJ20070460
- ↑ Boulin T, Rapti G, Briseno-Roa L, Stigloher C, Richmond JE, Paoletti P, Bessereau JL. Positive modulation of a Cys-loop acetylcholine receptor by an auxiliary transmembrane subunit. Nat Neurosci. 2012 Oct;15(10):1374-81. doi: 10.1038/nn.3197. Epub 2012 Aug 26. PMID:22922783 doi:http://dx.doi.org/10.1038/nn.3197
- ↑ Aran M, Smal C, Pellizza L, Gallo M, Otero LH, Klinke S, Goldbaum FA, Ithurralde ER, Bercovich A, Mac Cormack WP, Turjanski AG, Cicero DO. Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain. Proteins. 2014 Aug 13. doi: 10.1002/prot.24667. PMID:25116514 doi:http://dx.doi.org/10.1002/prot.24667
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Martin Aran, Alexander Berchansky, Joel L. Sussman