|
|
| (28 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | [[Image:1akz.png|left|200px|thumb|Crystal Structure of DNA glycosylate, [[1akz]]]]
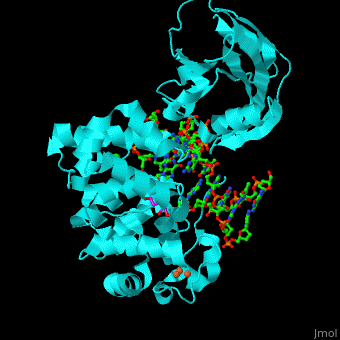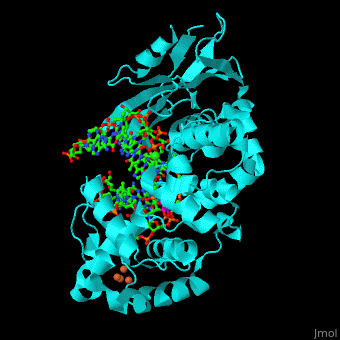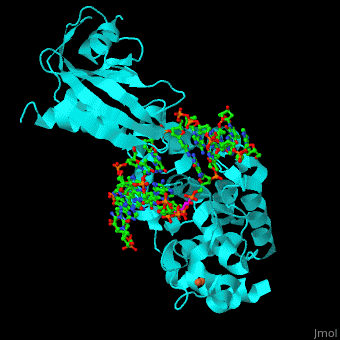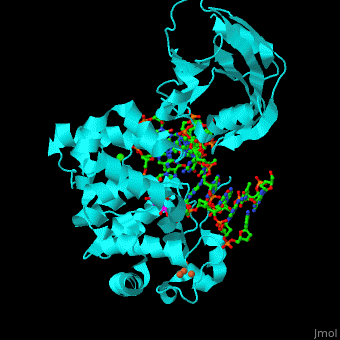
| + | <StructureSection load='' size='340' side='right' caption='Structure of adenine DNA glycosylase containing Fe4S4 cluster complex with DNA containing an oxoG base, deoxy-ribofuranose phosphate and Ca+2 ion (green) (PDB code [[1rrs]]).' scene='44/445369/Cv/1'> |
| - | {{STRUCTURE_3fsp| PDB=3fsp | SIZE=400| SCENE= |right|CAPTION=Adenine DNA glycosylate complex with DNA, Fe4-S4 cluster, phosphoric acid esther and Ca+2 ion [[3fsp]] }}
| + | __TOC__ |
| | + | == Function == |
| | | | |
| - | | + | '''DNA glycosylase''' (DG) are enzymes which remove damaged DNA bases by flipping them out of the double helix followed by their cleavage. Monofunctional DG have only glycosylase activity; bifunctional DG also act as lysates; ADG, UDG, TDG remove adenine, uracil, thymine from DNA.<ref>PMID:15102448</ref><br /> |
| - | | + | * '''Adenine DNA glycosylase''' is also called '''MutY'''. MutY has a role in prevention of DNA mutations resulting from oxidative damage forming the mutated base oxoG. DNA polymerase misreads oxoG and pairs it with adenine instead of cytosine. MutY removes adenine from the mismatched oxoG:A pair. MutY contains a 4Fe4S cluster.<br /> |
| - | | + | * '''Uracil DNA glycosylase''' (UDG) eliminates uracil from DNA and thus preventing mutagenesis. UDG initiates the base excision repair pathway. UDG excises uracil in both AU and GU pairs. UDG can repair 10,000 damaged bases in the human cell per day<ref>PMID:9882380</ref>. See also [[DNA Repair]]. |
| - | | + | * '''Thymine DNA glycosylase''' removes thymine moieties from mismatched G/T, C/T and T/T.<br /> |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | '''DNA glycosylase''' (DG) are enzymes which remove damaged DNA bases by flipping them out of the double helix followed by their cleavage. Monofunctional DG have only glycosylase activity; bifunctional DG also act as lysates; ADG, UDG, TDG remove adenine, uracil, thymine from DNA.<br /> | + | |
| - | * '''Adenine DNA glycosylase''' is also called '''MutY'''.<br /> | + | |
| | * '''Smug1''' is a single-strand selective monofunctional UDG. <br /> | | * '''Smug1''' is a single-strand selective monofunctional UDG. <br /> |
| | * '''8-oxoguanine DNA glycosylase''' recognize oxidized bases.<br /> | | * '''8-oxoguanine DNA glycosylase''' recognize oxidized bases.<br /> |
| Line 32: |
Line 15: |
| | *[[Fpg Nei Protein Superfamily]]. | | *[[Fpg Nei Protein Superfamily]]. |
| | | | |
| - | == 3D Structures of DNA glycosylate == | + | == Structural highlights == |
| - | | + | |
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}}
| + | |
| - | {{#tree:id=OrganizedByTopic|openlevels=0|
| + | |
| - | | + | |
| - | *Adenine DG
| + | |
| - | | + | |
| - | **[[3n5n]] – hADG catalytic domain – human<BR />
| + | |
| - | **[[1yuo]] - hADG catalytic domain (mutant) <BR />
| + | |
| - | **[[1x51]] – hADG NUDIX domain - NMR<BR />
| + | |
| - | **[[1wef]], [[1weg]], [[1kg4]], [[1kg5]], [[1kg6]], [[1kg7]], [[1kqj]], [[1mun]] - EcADG catalytic domain (mutant) - ''Escherichia coli''<BR />
| + | |
| - | **[[1kg2]], [[1kg3]], [[1muy]] - EcADG catalytic domain<BR />
| + | |
| - | **[[1mun]] - EcADG catalytic domain (mutant) <BR />
| + | |
| - | | + | |
| - | *Adenine DG complex
| + | |
| - | | + | |
| - | **[[1wei]], [[1mud]] - EcADG catalytic domain (mutant) + adenine<br />
| + | |
| - | **[[3fsp]], [[3fsq]] – GsADG + DNA – ''Geobacillus stearothermophilus''<BR />
| + | |
| - | **[[3g0q]], [[1vrl]], [[1rrq]], [[1rrs]] - GsADG (mutant) + DNA <BR />
| + | |
| - | | + | |
| - | *Uracil DG
| + | |
| - | | + | |
| - | **[[3a7n]] – MtUDG – ''Mycobacterium tuberculosis''<BR />
| + | |
| - | **[[3tr7]] – UDG – ''Coxiella burnetii''<br />
| + | |
| - | **[[1akz]], [[3tkb]] - hUDG (mutant) <BR />
| + | |
| - | **[[2jhq]] – UDG – ''Vibrio cholerae''<BR />
| + | |
| - | **[[2owq]], [[2owr]], [[3nt7]], [[4dof]], [[4dog]], [[4rib]], [[4lzb]] – UDG (mutant) – Vaccinia virus<BR />
| + | |
| - | **[[2d3y]], [[1ui0]], [[1ui1]] – TtUDG – ''Thermus thermophilus''<BR />
| + | |
| - | **[[1lqj]] – EcUDG<BR />
| + | |
| - | **[[1flz]] - EcUDG (mutant) <BR />
| + | |
| - | **[[1udh]], [[1udg]] – HsUDG - Herpes simplex virus<BR />
| + | |
| - | **[[2c2p]], [[2boo]] – DrUDG – ''Deinococcus radiodurans''<BR />
| + | |
| - | **[[2c2q]] – DrUDG (mutant) <BR />
| + | |
| - | **[[1vk2]], [[1l9g]] – TmUDG - ''Thermotoga maritima''<BR />
| + | |
| - | **[[1okb]] – AcUDG catalytic domain – Atlantic cod<br />
| + | |
| - | **[[3wdf]] – SaUDG - ''Staphylococcus aureus''<br />
| + | |
| - | **[[3zor]] – BsUDG – ''Bacillus subtilis''<br />
| + | |
| - | | + | |
| - | *Uracil DG complex
| + | |
| - | | + | |
| - | **[[2zhx]] – MtDG + protein inhibitor<BR />
| + | |
| - | **[[2oxm]], [[2oyt]], [[1q3f]] – hUDG + DNA<BR />
| + | |
| - | **[[1ssp]], [[2ssp]] - hUDG + DNA+ uracil<BR />
| + | |
| - | **[[1emh]], [[1emj]], [[4skn]] – hUDG (mutant) + DNA<BR />
| + | |
| - | **[[3fcf]], [[3fci]], [[3fck]], [[3fcl]], [[2hxm]] – hUDG + inhibitor<BR />
| + | |
| - | **[[1ugh]] - hUDG (mutant) + protein inhibitor<BR />
| + | |
| - | **[[3cxm]] – UDG residues 129-373 + 5-bromo-uracil<BR />
| + | |
| - | **[[2dp6]], [[2dem]], [[2ddg]] – TtUDG + DNA<BR />
| + | |
| - | **[[2j8x]] – UDG glycosylase domain + protein inhibitor – Epstein-Barr virus<BR />
| + | |
| - | **[[2eug]], [[3eug]], [[1eug]], [[4eug]], [[5eug]] - EcUDG (mutant) + uracil<BR />
| + | |
| - | **[[1mwj]], [[1mwi]], [[1mtl]] – EcUDG + DNA<BR />
| + | |
| - | **[[1lqg]], [[1lqm]], [[1eui]], [[1uug]] - EcUDG + protein inhibitor<BR />
| + | |
| - | **[[2uug]] - EcUDG (mutant) + protein inhibitor<BR />
| + | |
| - | **[[3uf7]] - EcUDG + single-stranded DNA-binding protein peptide<br />
| + | |
| - | **[[3ufm]] - DrUDG + single-stranded DNA-binding protein peptide<br />
| + | |
| - | **[[2c53]], [[2c56]] – HhUDG (mutant) – Human herpesvirus<BR />
| + | |
| - | **[[1lau]] – HhUDG + DNA<BR />
| + | |
| - | **[[1udi]] - HsUDG + protein inhibitor<BR />
| + | |
| - | **[[3wdg]] – SaUDG + protein<br />
| + | |
| - | **[[3zoq]] – BsUDG + p56 <br />
| + | |
| - | **[[4l5n]] – HsUDG + p56 <br />
| + | |
| - | **[[4lyl]] - AcUDG + protein inhibitor<br />
| + | |
| - | **[[4od8]], [[4oda]] - VvUDG + DNA polymerase processivity factor<br />
| + | |
| - | | + | |
| - | *Smug1 Uracil DG
| + | |
| - | | + | |
| - | **[[1oe4]], [[1oe5]], [[1oe6]] – UDG Smug1 + DNA– ''Xenopus laevis''
| + | |
| - | | + | |
| - | *Thymine DG (G/T mismatch-specific thymine DG)
| + | |
| - | | + | |
| - | **[[2rba]] – hTDG core domain + DNA<BR />
| + | |
| - | **[[2d07]], [[1wyw]] – hTDG central region + SUMO-3<BR />
| + | |
| - | **[[1kea]] – TDG – ''Methanothermobacter thermautotrophicus''<BR />
| + | |
| - | **[[1mug]] – EcTDG
| + | |
| - | **[[3uo7]], [[4jgc]] - hTDG + DNA + carboxylcytosine<br />
| + | |
| - | **[[3uob]] - hTDG + DNA + fluoro-cytidine<br />
| + | |
| - | **[[3ufj]] – hTDG core domain + DNA + fluoroluridine<br />
| + | |
| - | **[[4fnc]] - hTDG + DNA + hydroxymethyluracil<br />
| + | |
| - | | + | |
| - | *Human 8-oxoguanine DG
| + | |
| - | | + | |
| - | **[[1ko9]] – hOgg1<BR />
| + | |
| - | **[[2xhi]], [[3ktu]], [[1hu0]], [[1fn7]] - hOgg1 + DNA<BR />
| + | |
| - | **[[1lwv]], [[1lww]], [[1lwy]] - hOgg1 + DNA<BR /> + guanine derivative<BR />
| + | |
| - | **[[2nob]], [[2noe]], [[2nof]], [[2noh]], [[2noi]], [[2nol]], [[2noz]], [[2i5w]], [[1yqk]], [[1yql]], [[1yqm]], [[1yqr]], [[1n39]], [[1n3a]], [[1n3c]] - hOgg1 (mutant) + DNA<BR />
| + | |
| - | **[[1m3h]], [[3ih7]] - hOgg1 core fragment (mutant) + DNA<BR />
| + | |
| - | **[[1m3q]], [[1ebm]] - hOgg1 core fragment (mutant) + DNA + guanine derivative
| + | |
| - | | + | |
| - | *8-oxoguanine (oxoG) DG
| + | |
| - | | + | |
| - | **[[3n0u]] – TmOxDG<BR />
| + | |
| - | **[[3knt]] - MjOxDG (mutant) + DNA – ''Methanocaldococcus jannaschii''<BR />
| + | |
| - | **[[3fhf]] - MjOxDG<BR />
| + | |
| - | **[[3go8]], [[3gp1]], [[3gpp]], [[3gpu]], [[3gpx]], [[3gpy]], [[3gq3]], [[3gq4]], [[3gq5]] – GsOxDG (mutant) + DNA<BR />
| + | |
| - | **[[3f0z]] - CaOxDG – ''Clostridium acetobutylicum''<BR />
| + | |
| - | **[[3f10]] – CaOxDG + oxoG<BR />
| + | |
| - | **[[3i0w]], [[3i0x]] - CaOxDG (mutant) + DNA<BR />
| + | |
| - | **[[3fhg]] - OxDG (mutant) – ''Sulfolobus solfataricus''<BR />
| + | |
| - | **[[1xqo]], [[1xqp]] – OxDG – ''Pyrobaculum aerophilum''<br />
| + | |
| - | **[[4ejy]], [[4ejz]] – OxDG + DNA – ''Thermoanaerobacter tengcongensis''<br />
| + | |
| - | | + | |
| - | *Methyladenine DG
| + | |
| - | | + | |
| - | **[[3bvs]] – BcAlkDG – ''Bacillus cereus''<BR />
| + | |
| - | **[[1pvs]], [[1p7m]], [[1mpg]] – EcAlkDG<BR />
| + | |
| - | **[[1nku]], [[1lmz]] – EcAlkDG - NMR<BR />
| + | |
| - | **[[2ofk]] – StAlkDG – ''Salmonella typhi''<BR />
| + | |
| - | **[[2jhj]], [[2jhn]] – AlkDG – ''Archaeoglobus fulgidus''<BR />
| + | |
| - | **[[2jg6]] – AlkDG – ''Staphylococcus aureus''<BR />
| + | |
| - | **[[1pu6]] – HpAlkDG – ''Helicobacter pylori''<BR />
| + | |
| - | **[[4ai4]] – SaAlkDG (mutant)<br />
| + | |
| - | | + | |
| - | *Methyladenine DG complex
| + | |
| - | | + | |
| - | **[[3qi5]], [[1ewn]], [[1f4r]], [[1f6o]], [[1bnk]], [[3uby]] – hAlkDG + DNA<BR />
| + | |
| - | **[[3jx7]], [[3jxy]], [[3jxz]], [[3jy1]] - BcAlkDG + DNA<BR />
| + | |
| - | **[[3ogd]], [[3oh6]], [[3oh9]] - EcAlkDG (mutant) + DNA<BR />
| + | |
| - | **[[3d4v]], [[3cvs]], [[3cvt]], [[3cw7]], [[3cwa]], [[3cws]], [[3cwt]], [[3cwu]], [[1diz]] - EcAlkDG + DNA<BR />
| + | |
| - | **[[2ofi]] – StAlkDG + DNA<BR />
| + | |
| - | **[[3s6i]] – AlkDG + DNA – fission yeast<br />
| + | |
| - | **[[1pu7]], [[1pu8]] – HpAlkDG + adenine derivative<br />
| + | |
| - | **[[4aia]] – SaAlkDG + methyladenine<br />
| + | |
| - | **[[4ai5]] – SaAlkDG (mutant) + methyladenine<br />
| + | |
| - | | + | |
| - | *Formamidopyrimidine DG
| + | |
| - | | + | |
| - | **[[1ee8]] – TtFpg <br />
| + | |
| - | **[[3a42]], [[3a45]] – ApFpg – ''Acanthamoeba polyphaga'' minivirus<br />
| + | |
| - | **[[3twk]], [[3twl]] – AtFpg - ''Arabidopsis thaliana''<br />
| + | |
| - | | + | |
| - | *''Fpg complex with DNA''
| + | |
| - | | + | |
| - | **[[1k82]] – EcFpg + DNA <br />
| + | |
| - | **[[1kfv]], [[1nnj]] – LlFpg (mutant) + DNA – ''Lactococcus lactis''<br />
| + | |
| - | **[[1l1t]], [[1l1z]], [[1l2b]], [[1l2c]] – GsFpg + DNA <br />
| + | |
| - | **[[2f5n]], [[2f5o]], [[2f5p]], [[3gpx]], [[3gq5]] – GsFpg (mutant) + DNA <br />
| + | |
| | | | |
| - | *''Fpg complex with DNA containing a damaged base'' | + | MutY uses base flipping to twist the mispaired adenine out of the DNA helix and into the MutY active site. MutY contains 2 domains: C-terminal domain and catalytic domain containing a helix-hairpin-helix. |
| | + | *<scene name='44/445369/Cv/9'>Adenine DNA glycosylase containing Fe4S4 cluster with DNA containing an mispaired adenine 8OG and Ca+2</scene>. |
| | + | *<scene name='44/445369/Cv/10'>Mispaired adenine 8OG binding site</scene>. |
| | + | *<scene name='44/445369/Cv/11'>Fe4S4 cluster binds to 3 Cys residues</scene>. |
| | + | *<scene name='44/445369/Cv/12'>Ca+2 ion coordination site</scene> (PDB code [[1rrs]])<ref>PMID:14961129</ref>. Water molecules shown as red spheres. |
| | | | |
| - | **[[1r2y]], [[2f5q]], [[2f5s]], [[3jr5]], [[3gp1]], [[3gpp]], [[3gpu]], **[[3gpy]], [[3gq3]], [[3gq4]], [[3go8]], [[3u6c]], [[3u6d]] – GsFpg (mutant) + DNA containing oxoG<br />
| + | == 3D Structures of DNA glycosylase == |
| - | **[[3u6e]], [[4g4o]], [[4g4r]] – GsFpg (mutant) + DNA containing oxoG and thymidine derivative <br />
| + | [[DNA glycosylase 3D structures]] |
| - | **[[3u6l]], [[3u6m]], [[3u6q]] – GsFpg (mutant) + DNA containing oxoG and cytidine derivative <br />
| + | |
| - | **[[1r2z]] – GsFpg (mutant) + DNA containing 5,6-dihydrouracil<br />
| + | |
| - | **[[3saw]] – GsFpg + DNA containing cytidine derivative<br />
| + | |
| - | **[[3sar]], [[3sav]], [[3sbj]] – GsFpg (mutant) + DNA containing cytidine derivative<br />
| + | |
| - | **[[1l2d]], [[3sau]], [[3u6o]], [[3u6p]], [[3u6s]], [[4g4n]], [[4g4q]] – GsFpg (mutant) + DNA containing thymidine derivative<br />
| + | |
| - | **[[3sas]], [[3sat]] – GsFpg (mutant) + DNA containing open deoxy-ribofuranose phosphate <br />
| + | |
| - | **[[1pm5]] – LlFpg + DNA containing abasic dideoxyribose<br />
| + | |
| - | **[[1pjj]] – LlFpg (mutant) + DNA containing abasic dideoxyribose<br />
| + | |
| - | **[[4cis]] – LlFpg + DNA containing oxoG derivative<br />
| + | |
| - | **[[3twm]] – AtFpg + DNA containing abasic dideoxyribose<br />
| + | |
| - | **[[1pji]] – LlFpg + DNA containing 1,3-propanediol<br />
| + | |
| - | **[[2xzf]], [[2xzu]] – LlFpg + DNA containing oxidized pyrimidine<br />
| + | |
| - | **[[1tdz]], [[1xc8]], [[3c58]] – LlFpg + DNA containing Fapy-dG derivative<br />
| + | |
| - | **[[3a46]] – ApFpg + DNA containing abasic dideoxyribose<br />
| + | |
| - | **[[4nrw]] – ApFpg (mutant) + DNA containing abasic dideoxyribose<br />
| + | |
| - | **[[3vk7]] – ApFpg (mutant) + DNA containing 5-hydroxyuracil <br />
| + | |
| - | **[[3vk8]] – ApFpg (mutant) + DNA containing thymine glycol<br />
| + | |
| - | **[[3jr4]] – GsFpg (mutant) + DNA + guanosine derivative <br />
| + | |
| | | | |
| - | *DNA glycosylase NEI
| + | </StructureSection> |
| | | | |
| - | **[[2ea0]], [[2opf]], [[2oq4]], [[1k3w]], [[1k3x]] – EcGD NEI + DNA<BR />
| + | == References == |
| - | **[[2fcc]] – Pyrimidine GD + DNA – Enterobacteria phage T4<br />
| + | <references/> |
| - | **[[1q39]] – EcGD NEI<br />
| + | [[Category:Topic Page]] |
| - | **[[1q3b]], [[1q3c]] – EcGD NEI (mutant)<br />
| + | |
| - | **[[1tdh]] – hGD NEI
| + | |
| - | }}
| + | |