Calmodulin
From Proteopedia
(Difference between revisions)
| (32 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
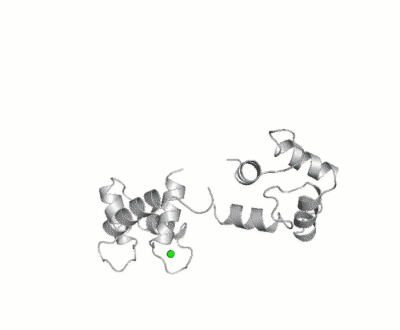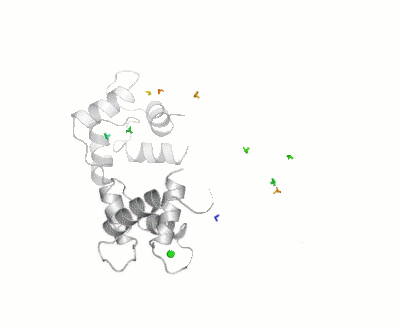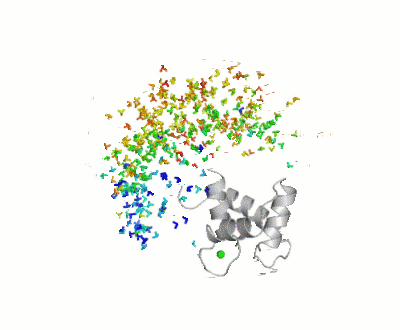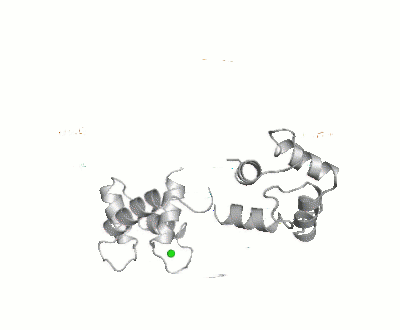
| - | + | <StructureSection load='1cll' size='350' side='right' scene='39/398280/Cv/3' caption='Human calmodulin complex with ethanol and Ca+2 ions (green) (PDB code [[1cll]])'> | |
| - | + | __TOC__ | |
| + | ==Function== | ||
| - | + | [[Calmodulin]] (CaM) – calcium modulated protein – regulates various protein targets. It is used by various proteins as calcium sensor and signal transducer by binding to their calcium binding domain (CBD). It undergoes conformational change upon binding Ca++ via its 4 [[EF hand]] motives and can undergo post-translational modification. <scene name='39/398280/Cv/4'>Click here to see EF hand</scene> of Human calmodulin (PDB code [[1cll]]). <ref>PMID:1474585</ref> More details on apo-CaM [[Calcium-free Calmodulin]] and [[Calmodulin JMU]]. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Calmodulin]] (CaM) – calcium modulated protein – regulates various protein targets. It is used by various proteins as calcium sensor and signal transducer by binding to their calcium binding domain (CBD). It undergoes conformational change upon binding Ca++ via its 4 [[EF hand]] motives and can undergo post-translational modification. | + | |
| - | + | ||
| - | + | ||
| - | + | ||
{{Clear}} | {{Clear}} | ||
| Line 38: | Line 15: | ||
== Calmodulin in Motion == | == Calmodulin in Motion == | ||
| - | The | + | The buttons below allow you to explore morphs <ref>The [[Jmol/Storymorph|Storymorph Jmol scripts]] creates the interpolated coordinates of the morph on the fly.</ref> between structures [[1prw]] and [[1cll]]. |
| - | < | + | <jmol> |
| + | <jmolButton> | ||
| + | <script> | ||
| + | script "/wiki/images/a/a2/Storymorph.spt"; | ||
| + | load files "=1prw" "=1cll"; | ||
| + | delete water; | ||
| + | delete protein and not backbone; | ||
| + | select all;cartoon only;cartoon off; | ||
| + | select 4-147;cartoon on; | ||
| + | model 1; | ||
| + | center visible;color group; | ||
| + | compare {2.1} {1.1} SUBSET{*.CA} ATOMS{4-138}{4-138} ROTATE TRANSLATE; | ||
| + | </script> | ||
| + | <text>Prepare Animation</text> | ||
| + | </jmolButton> | ||
| + | </jmol><jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | model 2; | ||
| + | display 4-147; | ||
| + | backbone only; | ||
| + | backbone 0.5 | ||
| + | structures = [{1.1}, {2.1}]; | ||
| + | my_recipe = [ | ||
| + | [{79-147},{79-147}, {(79-138) and alpha}], | ||
| + | [{4-78}, {79}, {(8-78) and alpha}], | ||
| + | ]; | ||
| + | morph(20, structures, my_recipe); | ||
| + | </script> | ||
| + | <text>Open</text> | ||
| + | </jmolButton> | ||
| + | </jmol><jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | model 1 | ||
| + | display 4-147; | ||
| + | backbone only; | ||
| + | backbone 0.5 | ||
| + | structures = [{2.1}, {1.1}]; | ||
| + | my_recipe = [ | ||
| + | [{79-147},{79-147}, {(79-138) and alpha}], | ||
| + | [{4-78}, {79}, {(8-78) and alpha}], | ||
| + | ]; | ||
| + | morph(20, structures, my_recipe); | ||
| + | </script> | ||
| + | <text>Close</text> | ||
| + | </jmolButton> | ||
| + | </jmol> | ||
| - | + | The following morph is between [[1prw]] and [[1cll]] after superposition of residues 79-138. This shows the subtle conformational changes in that domain more clearly. | |
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | script "/wiki/images/a/a2/Storymorph.spt"; | ||
| + | load files "=1prw" "=1cll"; | ||
| + | delete water; | ||
| + | delete protein and not backbone; | ||
| + | select all;cartoon only;cartoon off; | ||
| + | select 4-147;cartoon on; | ||
| + | model 1; | ||
| + | center visible;color group; | ||
| + | compare {2.1} {1.1} SUBSET{*.CA} ATOMS{79-138}{79-138} ROTATE TRANSLATE; | ||
| + | </script> | ||
| + | <text>Prepare Animation</text> | ||
| + | </jmolButton> | ||
| + | </jmol><jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | model 2; | ||
| + | display 4-147; | ||
| + | backbone only; | ||
| + | backbone 0.5 | ||
| + | structures = [{1.1}, {2.1}]; | ||
| + | my_recipe = [ | ||
| + | [{79-147},{79-147}, {(79-138) and alpha}], | ||
| + | [{4-78}, {79}, {(8-78) and alpha}], | ||
| + | ]; | ||
| + | morph(20, structures, my_recipe); | ||
| + | </script> | ||
| + | <text>Open</text> | ||
| + | </jmolButton> | ||
| + | </jmol><jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | model 1 | ||
| + | display 4-147; | ||
| + | backbone only; | ||
| + | backbone 0.5 | ||
| + | structures = [{2.1}, {1.1}]; | ||
| + | my_recipe = [ | ||
| + | [{79-147},{79-147}, {(79-138) and alpha}], | ||
| + | [{4-78}, {79}, {(8-78) and alpha}], | ||
| + | ]; | ||
| + | morph(20, structures, my_recipe); | ||
| + | </script> | ||
| + | <text>Close</text> | ||
| + | </jmolButton> | ||
| + | </jmol> | ||
| - | + | The following morph is between [[1prw]] and [[1cll]] after superposition of residues 8-78. This shows the subtle conformational changes in that domain more clearly. | |
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | script "/wiki/images/a/a2/Storymorph.spt"; | ||
| + | load files "=1prw" "=1cll"; | ||
| + | delete water; | ||
| + | delete protein and not backbone; | ||
| + | select all;cartoon only;cartoon off; | ||
| + | select 4-147;cartoon on; | ||
| + | model 1; | ||
| + | center visible;color group; | ||
| + | compare {2.1} {1.1} SUBSET{*.CA} ATOMS{8-78}{8-78} ROTATE TRANSLATE; | ||
| + | </script> | ||
| + | <text>Prepare Animation</text> | ||
| + | </jmolButton> | ||
| + | </jmol><jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | model 2; | ||
| + | display 4-147; | ||
| + | backbone only; | ||
| + | backbone 0.5 | ||
| + | structures = [{1.1}, {2.1}]; | ||
| + | my_recipe = [ | ||
| + | [{4-78}, {4-78}, {(8-78) and alpha}], | ||
| + | [{79-147},{78}, {(79-138) and alpha}], | ||
| + | ]; | ||
| + | morph(20, structures, my_recipe); | ||
| + | </script> | ||
| + | <text>Open</text> | ||
| + | </jmolButton> | ||
| + | </jmol><jmol> | ||
| + | <jmolButton> | ||
| + | <script> | ||
| + | model 1 | ||
| + | display 4-147; | ||
| + | backbone only; | ||
| + | backbone 0.5 | ||
| + | structures = [{2.1}, {1.1}]; | ||
| + | my_recipe = [ | ||
| + | [{4-78}, {4-78}, {(8-78) and alpha}], | ||
| + | [{79-147},{78}, {(79-138) and alpha}], | ||
| + | ]; | ||
| + | morph(20, structures, my_recipe); | ||
| + | </script> | ||
| + | <text>Close</text> | ||
| + | </jmolButton> | ||
| + | </jmol> | ||
| - | This movie was created by Andrei, Zini et al., of the | ||
| - | [http://www.scivis.ifc.cnr.it Scientific Visualization Unit], | ||
| - | Institute of Clinical Physiology - CNR of Itlay. | ||
| - | [http://www.molmovdb.org/cgi-bin/morph.cgi?ID=b097743-28520 Conformational change of Calmodulin] | ||
| - | |||
| - | <applet load='1cll' size='350' color='white' frame='true' align='right' caption='Calmodulin showing Ca+2 ions' scene='Calmodulin/Inicio/1' /> | ||
| - | |||
| - | <scene name='Calmodulin/Inicio/1'>Unbound</scene>, | ||
| - | <scene name='Calmodulin/Bound/4'>Bound</scene> | ||
| - | |||
| - | {{Clear}} | ||
== 3D Structures of Calmodulin == | == 3D Structures of Calmodulin == | ||
| + | [[Calmodulin 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==See Also== | ==See Also== | ||
Current revision
| |||||||||||
See Also
Bibliography
- ↑ Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. 1992 Dec 20;228(4):1177-92. PMID:1474585
- ↑ Bertini I, Giachetti A, Luchinat C, Parigi G, Petoukhov MV, Pierattelli R, Ravera E, Svergun DI. Conformational Space of Flexible Biological Macromolecules from Average Data. J Am Chem Soc. 2010 Sep 7. PMID:20822180 doi:10.1021/ja1063923
- ↑ The Storymorph Jmol scripts creates the interpolated coordinates of the morph on the fly.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Karsten Theis, Jaime Prilusky, Enrico Ravera, Daniel Moyano-Marino