|
|
| (47 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | number of documents: 595
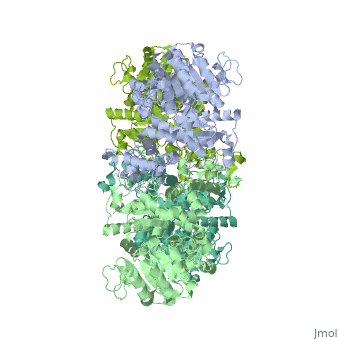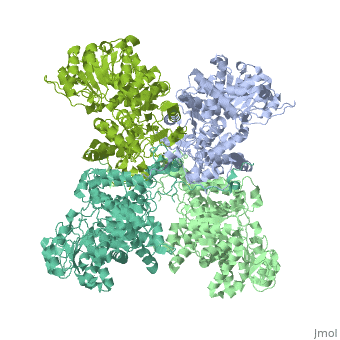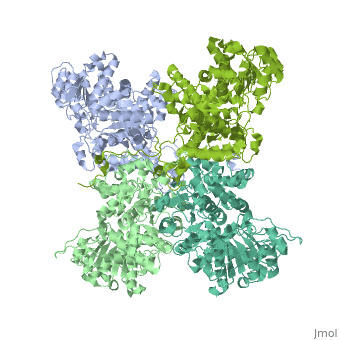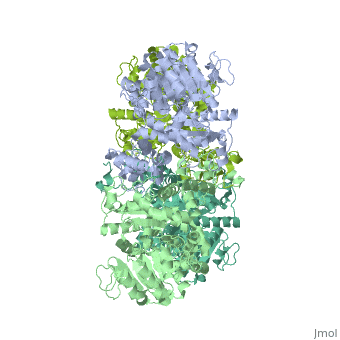
| + | <StructureSection load='3wja' size='340' side='right' caption='The best structure for NADP-dependent malic enzyme shown: [[3wja]]' scene=''> |
| - | == Lysozyme ==
| + | Best example is PDB entry [[3wja]] and is shown in the viewer.<br> |
| - | <StructureSection load='1swy' size='340' side='right' caption='Caption for this structure' scene=''> | + | ==Catalytic Activity == |
| - | Best example is 1swy<br> | + | Oxaloacetate = pyruvate + CO(2).Data source: Uniprot [http://www.uniprot.org/uniprot/P48163 P48163]<br> |
| - | Molecule Lysozyme, also known as Lysozyme, CP-7 lysin, Muramidase, Endolysin, 1,4-beta-N-acetylmuramidase, Lysis protein, CP-1 lysin, Lysozyme murein hydrolase, Protein gp17, Transglycosylase, Hevamine-A, Chitinase and L-alanyl-D-glutamate peptidase.
| + | Or the decarboxylation of malate to pyruvate<ref>PMID:8187880</ref>. |
| - | == Function == | + | |
| - | lysozyme activity<br>
| + | |
| - | catalytic activity<br>
| + | |
| - | hydrolase activity<br>
| + | |
| - | hydrolase activity, acting on glycosyl bonds<br>
| + | |
| - | carbon-oxygen lyase activity, acting on polysaccharides<br>
| + | |
| - | lyase activity<br>
| + | |
| - | chitinase activity<br>
| + | |
| - | hydrolase activity, hydrolyzing O-glycosyl compounds<br>
| + | |
| - | sequence-specific DNA binding<br>
| + | |
| | | | |
| - | == Disease == | |
| - | | |
| - | == Relevance == | |
| - | | |
| - | == Structural highlights == | |
| | == Biological process == | | == Biological process == |
| | Is involved in the following biological processes:<br> | | Is involved in the following biological processes:<br> |
| - | cell wall macromolecule catabolic process<br>
| + | malate metabolic process<br> |
| - | peptidoglycan catabolic process<br>
| + | oxidation-reduction process<br> |
| - | metabolic process<br>
| + | response to carbohydrate<br> |
| - | defense response to bacterium<br>
| + | response to hormone<br> |
| - | cytolysis<br>
| + | |
| - | viral release from host cell<br>
| + | |
| | carbohydrate metabolic process<br> | | carbohydrate metabolic process<br> |
| - | chitin catabolic process<br>
| + | protein tetramerization<br> |
| - | polysaccharide catabolic process<br>
| + | regulation of NADP metabolic process<br> |
| - | cytolysis by virus of host cell<br>
| + | NADP biosynthetic process<br> |
| - | cell wall organization<br>
| + | small molecule metabolic process<br> |
| | + | cellular lipid metabolic process<br> |
| | == In structures == | | == In structures == |
| - | Lysozyme is found in 595 PDB entries<br>
| + | NADP-dependent malic enzyme is found in 3 PDB entries<br> |
| - | <br><b>Homo sapiens</b> (1 PDB entries): <br>
| + | *<b>Eukaryota</b> (3 PDB entries): <br> |
| - | [[4uis]] <div class="pdb-prints 4uis"></div><br>4.4 A resolution. Title: The cryoEM structure of human gamma-Secretase complex[http://www.ebi.ac.uk/pdbe/entry/emdb/emd-2974 emd-2974]. Other macromolecules also in this entry: GAMMA-SECRETASE, Nicastrin, Presenilin-1 NTF subunit, .<br>
| + | **<b>Homo sapiens</b> (2 PDB entries): <br> |
| - | <br><b>Enterobacteria phage P22</b> (2 PDB entries): <br>
| + | ***[[3wja]] <div class="pdb-prints 3wja"></div> |
| - | [[2anv]] <div class="pdb-prints 2anv"></div><br>1.04 A resolution. Title: crystal structure of P22 lysozyme mutant L86M<br> | + | ***Title: The crystal structure of human cytosolic NADP(+)-dependent malic enzyme in apo form |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Enterobacteria phage P22&molecule_name:Lysozyme] Search for other PDB entries<br><b>Enterobacteria phage P21</b> (2 PDB entries): <br>
| + | ***2.548 A resolution |
| - | [[3hdf]] <div class="pdb-prints 3hdf"></div><br>1.7 A resolution. Title: Crystal structure of truncated endolysin R21 from phage 21<br>
| + | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Homo%20sapiens%22&all_molecule_names:%22NADP-dependent%20malic%20enzyme%22&!chimera:y Search the PDB for NADP-dependent malic enzyme from Homo sapiens] |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Enterobacteria phage P21&molecule_name:Lysozyme] Search for other PDB entries<br><b>Hevea brasiliensis</b> (7 PDB entries): <br> | + | **<b>Columba livia</b> (1 PDB entries): <br> |
| - | [[1kqy]] <div class="pdb-prints 1kqy"></div><br>1.92 A resolution. Title: Hevamine Mutant D125A/E127A/Y183F in Complex with Penta-NAG<br>
| + | ***[[1gq2]] <div class="pdb-prints 1gq2"></div> |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Hevea brasiliensis&molecule_name:Lysozyme] Search for other PDB entries<br><b>Escherichia phage T5</b> (1 PDB entries): <br>
| + | ***Title: MALIC ENZYME FROM PIGEON LIVER |
| - | [[2mxz]] <div class="pdb-prints 2mxz"></div><br>N/A A resolution. Title: Bacteriophage T5 l-alanoyl-d-glutamate peptidase comlpex with Zn2+ (Endo T5-ZN2+)<br>
| + | ***2.5 A resolution |
| - | <br><b>Streptococcus phage CP-7</b> (1 PDB entries): <br>
| + | == Alternative names for NADP-dependent malic enzyme == |
| - | [[4cvd]] <div class="pdb-prints 4cvd"></div><br>1.666 A resolution. Title: Crystal structure of the central repeat of cell wall binding module of Cpl7<br> | + | Molecule '''NADP-dependent malic enzyme''', also known as '''NADP-ME''', '''NADP-dependent malic enzyme''' and '''Malic enzyme 1'''. |
| - | <br><b>Antheraea mylitta</b> (1 PDB entries): <br>
| + | |
| - | [[1iiz]] <div class="pdb-prints 1iiz"></div><br>2.4 A resolution. Title: Crystal Structure of the Induced Antibacterial Protein from Tasar Silkworm, Antheraea mylitta<br>
| + | == 3D Structures of NADP-dependent malic enzyme == |
| - | <br><b>Mus musculus</b> (1 PDB entries): <br>
| + | |
| - | [[1iak]] <div class="pdb-prints 1iak"></div><br>1.9 A resolution. Title: HISTOCOMPATIBILITY ANTIGEN I-AKOther macromolecules also in this entry: H-2 class II histocompatibility antigen, A-K beta chain, H-2 class II histocompatibility antigen, A-K alpha chain.<br>
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} |
| - | <br><b>Bombyx mori</b> (2 PDB entries): <br>
| + | |
| - | [[1gd6]] <div class="pdb-prints 1gd6"></div><br>2.5 A resolution. Title: STRUCTURE OF THE BOMBYX MORI LYSOZYME<br> | + | |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Bombyx mori&molecule_name:Lysozyme] Search for other PDB entries<br><b>Streptococcus phage Cp-1</b> (6 PDB entries): <br> | + | [[3wja]], [[2aw5]] – hME - human<br /> |
| - | [[2j8g]] <div class="pdb-prints 2j8g"></div><br>1.69 A resolution. Title: Crystal structure of the modular Cpl-1 endolysin complexed with a peptidoglycan analogue (E94Q mutant in complex with a tetrasaccharide- pentapeptide)<br> | + | [[7x11]], [[7x12]] – hME1 + NADP<br /> |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Streptococcus phage Cp-1&molecule_name:Lysozyme] Search for other PDB entries<br><b>Enterobacteria phage T4</b> (562 PDB entries): <br> | + | [[8e76]], [[8eyn]] – hME3<br /> |
| - | [[1swy]] <div class="pdb-prints 1swy"></div><br>1.06 A resolution. Title: Use of a Halide Binding Site to Bypass the 1000-atom Limit to Ab initio Structure Determination<br> | + | [[8e78]], [[8e8o]], [[8eyo]] – hME3 + NADP<br /> |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Enterobacteria phage T4&molecule_name:Lysozyme] Search for other PDB entries<br><b>Enterobacteria phage P1</b> (2 PDB entries): <br> | + | [[1gq2]] – ME + NADP - pigeon<br /> |
| - | [[1xju]] <div class="pdb-prints 1xju"></div><br>1.07 A resolution. Title: Crystal structure of secreted inactive form of P1 phage endolysin Lyz<br> | + | [[5ou5]] – ME - maize<br /> |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Enterobacteria phage P1&molecule_name:Lysozyme] Search for other PDB entries<br><b>Enterobacteria phage lambda</b> (3 PDB entries): <br> | + | |
| - | [[1am7]] <div class="pdb-prints 1am7"></div><br>2.3 A resolution. Title: Lysozyme from bacteriophage lambda<br> | + | == References == |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Enterobacteria phage lambda&molecule_name:Lysozyme] Search for other PDB entries<br><b>Meretrix lusoria</b> (3 PDB entries): <br>
| + | <references/> |
| - | [[4pj2]] <div class="pdb-prints 4pj2"></div><br>1.24 A resolution. Title: Crystal structure of Aeromonas hydrophila PliI in complex with Meretrix lusoria lysozymeOther macromolecules also in this entry: Putative exported protein, .<br> | + | [[Category:Topic Page]][[Category:PDBe]] |
| - | [http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific:Meretrix lusoria&molecule_name:Lysozyme] Search for other PDB entries<br><b>Ruditapes philippinarum</b> (1 PDB entries): <br>
| + | |
| - | [[2dqa]] <div class="pdb-prints 2dqa"></div><br>1.6 A resolution. Title: Crystal Structure of Tapes japonica Lysozyme<br> | + | |
| - | <br><b>Escherichia coli</b> (1 PDB entries): <br> | + | |
| - | [[2qb0]] <div class="pdb-prints 2qb0"></div><br>2.56 A resolution. Title: Structure of the 2TEL crystallization module fused to T4 lysozyme with an Ala-Gly-Pro linker.Other macromolecules also in this entry: Transcription factor ETV6, .<br> | + | |