Nitrile hydratase
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
'''Nitrile hydratase''' (NH) catalyze the hydration of nitriles to amides. NH contains Fe or Co ion in its active site. NH consists of two subunits: α and β and exists as dimer or tetramer with one metal ion bound per dimer. | '''Nitrile hydratase''' (NH) catalyze the hydration of nitriles to amides. NH contains Fe or Co ion in its active site. NH consists of two subunits: α and β and exists as dimer or tetramer with one metal ion bound per dimer. | ||
| - | |||
| - | == Disease == | ||
== Relevance == | == Relevance == | ||
| Line 13: | Line 11: | ||
== Structural highlights == | == Structural highlights == | ||
| - | === Crystal structure of the CN-hydrolase SA0302 from the pathogenic bacterium ''Staphylococcus aureus'' belonging to the Nit and NitFhit Branch of the nitrilase superfamily | + | === Crystal structure of the CN-hydrolase SA0302 from the pathogenic bacterium ''Staphylococcus aureus'' belonging to the Nit and NitFhit Branch of the nitrilase superfamily <ref>doi 10.1080/07391102.2012.719111</ref>=== |
| - | + | ||
| - | + | ||
| - | + | ||
The nitrilase, superfamily of protein enzyme, has in total over 180 known members. It includes a variety of thiol amidase enzymes involved in biosynthesis in plants, animals, fungi and prokaryotes <ref name="Pace">PMID:11380987</ref>. | The nitrilase, superfamily of protein enzyme, has in total over 180 known members. It includes a variety of thiol amidase enzymes involved in biosynthesis in plants, animals, fungi and prokaryotes <ref name="Pace">PMID:11380987</ref>. | ||
| Line 40: | Line 37: | ||
These data implicate the specificity of substrate binding for a definite member of Branch 10 of the Nit-nitrilase superfamily. The present solution of the substrate-free bacterial protein structure of SA0302 gives the basis for substrate binding studies with the potential substrates or inhibitors. | These data implicate the specificity of substrate binding for a definite member of Branch 10 of the Nit-nitrilase superfamily. The present solution of the substrate-free bacterial protein structure of SA0302 gives the basis for substrate binding studies with the potential substrates or inhibitors. | ||
| + | |||
| + | === Analyzing the Catalytic Role of Active Site Residues in the Fe-Type Nitrile Hydratase from ''Comamonas testosteroni'' Ni1 <ref>doi 10.1007/s00775-015-1273-3</ref>=== | ||
| + | |||
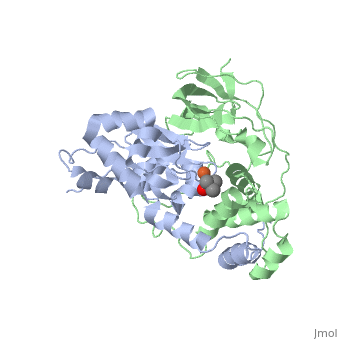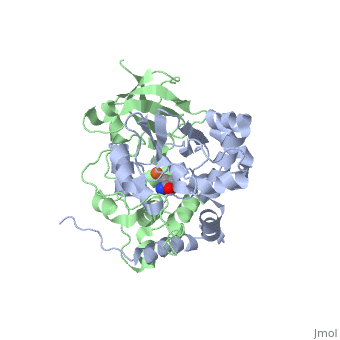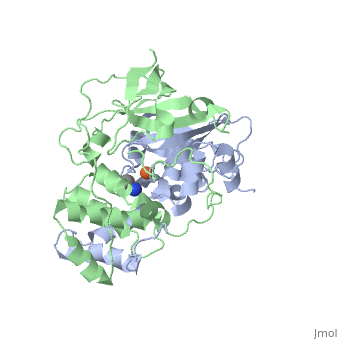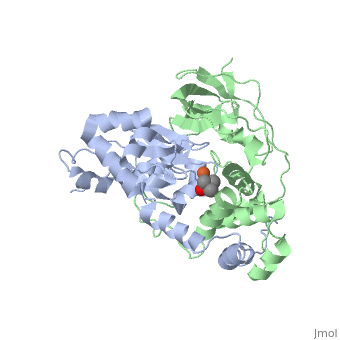
| + | <scene name='70/703434/Cv/3'>Active site of wild-type Fe-type nitrile hydratase</scene> from ''Comamonas testosteroni'' Ni1 (CtNHase, PDB ID: [[4fm4]]) in ball-and-stick form is shown. A strictly conserved active site arginine residue (αR157) and two histidine residues (αH80 and αH81) located near the active site of the CtNHase, <scene name='70/703434/Cv/8'>were mutated</scene>. <font color='darkmagenta'><b>The wild-type is in dark magenta</b></font>, <span style="color:lime;background-color:black;font-weight:bold;">αH80A/αH81A mutant is in green</span>, <span style="color:orange;background-color:black;font-weight:bold;">αH80W/αH81W is in orange</span>, and the <span style="color:deepskyblue;background-color:black;font-weight:bold;">αR157A mutant is in deep sky blue</span>. <scene name='70/703434/Cv/9'>Click here to see animation of this scene</scene>. These mutant enzymes were examined for their ability to bind iron and hydrate acrylonitrile. For the αR157A mutant, the residual activity (k<sub>cat</sub> = 10 ± 2 s−1) accounts for less than 1 % of the wild-type activity (k<sub>cat</sub> = 1100 ± 30 s<sup>-1</sup>) while the K<sub>m</sub> value is nearly unchanged at 205 ± 10 mM. On the other hand, mutation of the active site pocket αH80 and αH81 residues to alanine resulted in enzymes with k<sub>cat</sub> values of 220 ± 40 and 77 ± 13 s<sup>-1</sup>, respectively, and K<sub>m</sub> values of 187 ± 11 and 179 ± 18 mM. The double mutant (αH80A/αH81A) was also prepared and provided an enzyme with a k<sub>cat</sub> value of 132 ± 3 s<sup>-1</sup> and a K<sub>m</sub> value of 213 ± 61 mM. These data indicate that all three residues are catalytically important, but not essential. X-ray crystal structures of the <scene name='70/703434/Cv/11'>αH80A/αH81A</scene>, <scene name='70/703434/Cv/12'>αH80W/αH81W</scene>, and <scene name='70/703434/Cv/13'>αR157A</scene> mutant CtNHase enzymes were solved to 2.0, 2.8, and 2.5 Å resolutions, respectively. In each mutant enzyme, <scene name='70/703434/Cv/14'>hydrogen-bonding interactions crucial for the catalytic function of the αCys104-SOH ligand are disrupted</scene>. Disruption of these hydrogen bonding interactions likely alters the nucleophilicity of the sulfenic acid oxygen and the Lewis acidity of the active site Fe(III) ion. | ||
| + | <scene name='70/703434/Cv/4'>Superposition of the αβ heterodimer</scene>, the regions with pronounced difference between the wild-type and the mutant enzymes are indicated. <scene name='70/703434/Cv/7'>Click here to see animation of this scene</scene>. | ||
</StructureSection> | </StructureSection> | ||
| Line 51: | Line 53: | ||
**[[1ahj]], [[2cyz]], [[2cz0]], [[2cz1]] – ReNH α,β subunits – ''Rhodococcus erythropolis''<br /> | **[[1ahj]], [[2cyz]], [[2cz0]], [[2cz1]] – ReNH α,β subunits – ''Rhodococcus erythropolis''<br /> | ||
**[[2zcf]], [[3a8l]] – ReNH α (mutant),β subunits <br /> | **[[2zcf]], [[3a8l]] – ReNH α (mutant),β subunits <br /> | ||
| - | **[[4fm4]] – | + | **[[4fm4]], [[4zgd]] – CtNH α,β subunits – ''Comamonas testosteroni''<br /> |
| + | **[[4zge]], [[4zgj]] – CtNH α (mutant),β subunits <br /> | ||
| + | **[[6lk7]] – NH α subunit – ''Pseudomonas''<br /> | ||
*Fe-containing nitrile hydratase complex | *Fe-containing nitrile hydratase complex | ||
| Line 64: | Line 68: | ||
**[[3a8g]] – ReNH α (mutant),β subunits + trimethyl acetonitrile + NO <br /> | **[[3a8g]] – ReNH α (mutant),β subunits + trimethyl acetonitrile + NO <br /> | ||
**[[3a8h]] – ReNH α (mutant),β subunits + trimethyl acetonitrile <br /> | **[[3a8h]] – ReNH α (mutant),β subunits + trimethyl acetonitrile <br /> | ||
| - | **[[3a8m]] – ReNH α,β (mutant) subunits + trimethyl acetonitrile <br /> | + | **[[3a8m]], [[3wvd]], [[3wve]], [[3x20]], [[3x24]], [[3x25]], [[3x26]], [[3x28]] – ReNH α,β (mutant) subunits + trimethyl acetonitrile <br /> |
*'''Co-containing nitrile hydratase''' | *'''Co-containing nitrile hydratase''' | ||
**[[1ire]], [[1ugq]], [[4ob0]], [[4ob3]] – PtNH α,β subunits – ''Pseudonocardia thermophila''<br /> | **[[1ire]], [[1ugq]], [[4ob0]], [[4ob3]] – PtNH α,β subunits – ''Pseudonocardia thermophila''<br /> | ||
| - | **[[1ugr]], [[1ugs]], [[3vyh]] – PtNH α (mutant),β subunits <br /> | + | **[[1ugr]], [[1ugs]], [[3vyh]], [[7sjz]], [[7w8l]] – PtNH α (mutant),β subunits <br /> |
| + | **[[7qom]] – PtNH α,β (mutant) subunits <br /> | ||
| + | **[[7qom]], [[7qou]], [[7qoy]] – PtNH α (mutant),β (mutant) subunits <br /> | ||
**[[1v29]] – NH α,β subunits – ''Bacillus smithii''<br /> | **[[1v29]] – NH α,β subunits – ''Bacillus smithii''<br /> | ||
**[[2dpp]] – NH α,β subunits – ''Bacillus'' <br /> | **[[2dpp]] – NH α,β subunits – ''Bacillus'' <br /> | ||
| - | **[[3qxe]], [[3qyg]], [[3qz5]], [[3qz9]] – NH α,β subunits – ''Pseudomonas putida''<br /> | + | **[[3qxe]], [[3qyg]], [[3qyh]], [[3qz5]], [[3qz9]] – NH α,β subunits – ''Pseudomonas putida''<br /> |
| - | **[[ | + | **[[7qop]] – GpNH α (mutant),β subunits – ''Geobacillus palilidus''<br /> |
| + | **[[3hht]], [[7z0v]] – GpNH α (mutant),β (mutant) subunits <br /> | ||
**[[1ugp]] – PtNH α,β subunits + butyrate<br /> | **[[1ugp]] – PtNH α,β subunits + butyrate<br /> | ||
**[[4ob1]], [[4ob2]] – PtNH α,β subunits + butane boronic acid<br /> | **[[4ob1]], [[4ob2]] – PtNH α,β subunits + butane boronic acid<br /> | ||
Current revision
| |||||||||||
3D structures of nitrile hydratase
Updated on 18-July-2023
References
- ↑ Gordon RD, Qiu W, Romanov V, Lam K, Soloveychik M, Benetteraj D, Battaile KP, Chirgadze YN, Pai EF, Chirgadze NY. Crystal structure of the CN-hydrolase SA0302 from the pathogenic bacterium Staphylococcus aureus belonging to the Nit and NitFhit Branch of the nitrilase superfamily. J Biomol Struct Dyn. 2013 Oct;31(10):1057-65. doi: 10.1080/07391102.2012.719111. , Epub 2012 Oct 2. PMID:23607706 doi:http://dx.doi.org/10.1080/07391102.2012.719111
- ↑ 2.0 2.1 2.2 Pace HC, Brenner C. The nitrilase superfamily: classification, structure and function. Genome Biol. 2001;2(1):REVIEWS0001. Epub 2001 Jan 15. PMID:11380987
- ↑ Brenner C. Catalysis in the nitrilase superfamily. Curr Opin Struct Biol. 2002 Dec;12(6):775-82. PMID:12504683
- ↑ Kumaran D, Eswaramoorthy S, Gerchman SE, Kycia H, Studier FW, Swaminathan S. Crystal structure of a putative CN hydrolase from yeast. Proteins. 2003 Aug 1;52(2):283-91. PMID:12833551 doi:http://dx.doi.org/10.1002/prot.10417
- ↑ Barglow KT, Saikatendu KS, Bracey MH, Huey R, Morris GM, Olson AJ, Stevens RC, Cravatt BF. Functional Proteomic and Structural Insights into Molecular Recognition in the Nitrilase Family Enzymes (dagger) (double dagger). Biochemistry. 2008 Nov 24. PMID:19053248 doi:10.1021/bi801786y
- ↑ Martinez S, Wu R, Krzywda K, Opalka V, Chan H, Liu D, Holz RC. Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1. J Biol Inorg Chem. 2015 Jul;20(5):885-94. doi: 10.1007/s00775-015-1273-3. Epub, 2015 Jun 16. PMID:26077812 doi:http://dx.doi.org/10.1007/s00775-015-1273-3