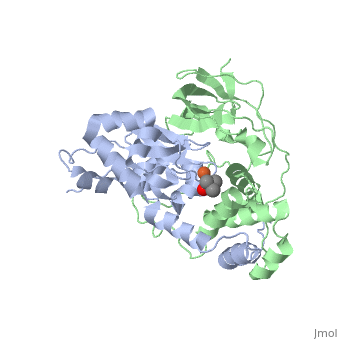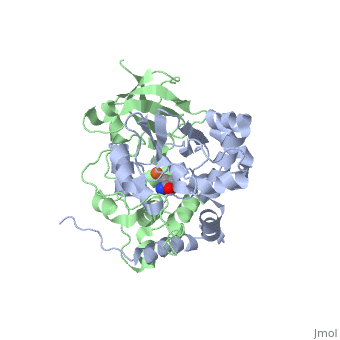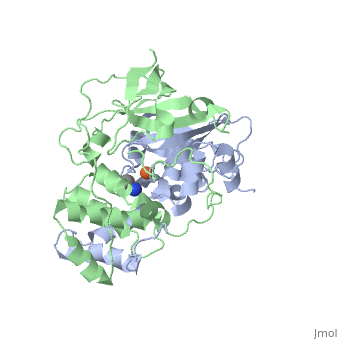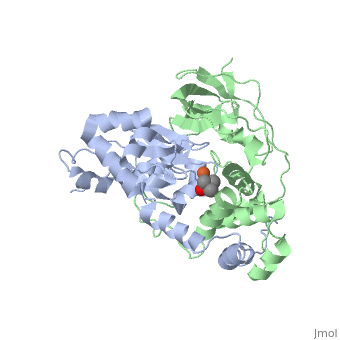
Nitrile hydratase
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 55: | Line 55: | ||
**[[4fm4]], [[4zgd]] – CtNH α,β subunits – ''Comamonas testosteroni''<br /> | **[[4fm4]], [[4zgd]] – CtNH α,β subunits – ''Comamonas testosteroni''<br /> | ||
**[[4zge]], [[4zgj]] – CtNH α (mutant),β subunits <br /> | **[[4zge]], [[4zgj]] – CtNH α (mutant),β subunits <br /> | ||
| + | **[[6lk7]] – NH α subunit – ''Pseudomonas''<br /> | ||
*Fe-containing nitrile hydratase complex | *Fe-containing nitrile hydratase complex | ||
| Line 72: | Line 73: | ||
**[[1ire]], [[1ugq]], [[4ob0]], [[4ob3]] – PtNH α,β subunits – ''Pseudonocardia thermophila''<br /> | **[[1ire]], [[1ugq]], [[4ob0]], [[4ob3]] – PtNH α,β subunits – ''Pseudonocardia thermophila''<br /> | ||
| - | **[[1ugr]], [[1ugs]], [[3vyh]] – PtNH α (mutant),β subunits <br /> | + | **[[1ugr]], [[1ugs]], [[3vyh]], [[7sjz]], [[7w8l]] – PtNH α (mutant),β subunits <br /> |
| + | **[[7qom]] – PtNH α,β (mutant) subunits <br /> | ||
| + | **[[7qom]], [[7qou]], [[7qoy]] – PtNH α (mutant),β (mutant) subunits <br /> | ||
**[[1v29]] – NH α,β subunits – ''Bacillus smithii''<br /> | **[[1v29]] – NH α,β subunits – ''Bacillus smithii''<br /> | ||
**[[2dpp]] – NH α,β subunits – ''Bacillus'' <br /> | **[[2dpp]] – NH α,β subunits – ''Bacillus'' <br /> | ||
| - | **[[3qxe]], [[3qyg]], [[3qz5]], [[3qz9]] – NH α,β subunits – ''Pseudomonas putida''<br /> | + | **[[3qxe]], [[3qyg]], [[3qyh]], [[3qz5]], [[3qz9]] – NH α,β subunits – ''Pseudomonas putida''<br /> |
| - | **[[ | + | **[[7qop]] – GpNH α (mutant),β subunits – ''Geobacillus palilidus''<br /> |
| + | **[[3hht]], [[7z0v]] – GpNH α (mutant),β (mutant) subunits <br /> | ||
**[[1ugp]] – PtNH α,β subunits + butyrate<br /> | **[[1ugp]] – PtNH α,β subunits + butyrate<br /> | ||
**[[4ob1]], [[4ob2]] – PtNH α,β subunits + butane boronic acid<br /> | **[[4ob1]], [[4ob2]] – PtNH α,β subunits + butane boronic acid<br /> | ||
Current revision
| |||||||||||
3D structures of nitrile hydratase
Updated on 18-July-2023
References
- ↑ Gordon RD, Qiu W, Romanov V, Lam K, Soloveychik M, Benetteraj D, Battaile KP, Chirgadze YN, Pai EF, Chirgadze NY. Crystal structure of the CN-hydrolase SA0302 from the pathogenic bacterium Staphylococcus aureus belonging to the Nit and NitFhit Branch of the nitrilase superfamily. J Biomol Struct Dyn. 2013 Oct;31(10):1057-65. doi: 10.1080/07391102.2012.719111. , Epub 2012 Oct 2. PMID:23607706 doi:http://dx.doi.org/10.1080/07391102.2012.719111
- ↑ 2.0 2.1 2.2 Pace HC, Brenner C. The nitrilase superfamily: classification, structure and function. Genome Biol. 2001;2(1):REVIEWS0001. Epub 2001 Jan 15. PMID:11380987
- ↑ Brenner C. Catalysis in the nitrilase superfamily. Curr Opin Struct Biol. 2002 Dec;12(6):775-82. PMID:12504683
- ↑ Kumaran D, Eswaramoorthy S, Gerchman SE, Kycia H, Studier FW, Swaminathan S. Crystal structure of a putative CN hydrolase from yeast. Proteins. 2003 Aug 1;52(2):283-91. PMID:12833551 doi:http://dx.doi.org/10.1002/prot.10417
- ↑ Barglow KT, Saikatendu KS, Bracey MH, Huey R, Morris GM, Olson AJ, Stevens RC, Cravatt BF. Functional Proteomic and Structural Insights into Molecular Recognition in the Nitrilase Family Enzymes (dagger) (double dagger). Biochemistry. 2008 Nov 24. PMID:19053248 doi:10.1021/bi801786y
- ↑ Martinez S, Wu R, Krzywda K, Opalka V, Chan H, Liu D, Holz RC. Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1. J Biol Inorg Chem. 2015 Jul;20(5):885-94. doi: 10.1007/s00775-015-1273-3. Epub, 2015 Jun 16. PMID:26077812 doi:http://dx.doi.org/10.1007/s00775-015-1273-3