Phosphoenolpyruvate carboxylase
From Proteopedia
| (3 intermediate revisions not shown.) | |||
| Line 20: | Line 20: | ||
== PEPC enzyme structure == | == PEPC enzyme structure == | ||
| - | Overall, plant PEPC’s present a similar structure characterized by four 105-110kDa identical subunits with a conserved N-terminal serine-phosphorylation domain, forming homotetramers <ref name="kai2003"/><ref name="O'Leary2011"/>. X-ray crystallography studies on ''Escherichia coli'' and maize (''Zea mays'') PEPC’s show that the tetramer is comprised of two pairs of monomers with a greater amount of intersubunit contacts, suggesting a homotetrameric structure of a ‘‘dimer-of-dimers’’. The dimers are held together through an interaction between Arg-438 of one subunit and Glu-433 of the neighboring subunit, forming a salt bridge <ref>PMID: 9927652</ref>. The monomeric structure maize’s C4-PEPC monomer consists of an eight-stranded β barrel and 42 α helices <ref name="kai2003"/><ref name="Izui2004">PMID: 15725057</ref>. The <scene name='57/573979/Cv/ | + | Overall, plant PEPC’s present a similar structure characterized by four 105-110kDa identical subunits with a conserved N-terminal serine-phosphorylation domain, forming homotetramers <ref name="kai2003"/><ref name="O'Leary2011"/>. X-ray crystallography studies on ''Escherichia coli'' and maize (''Zea mays'') PEPC’s show that the tetramer is comprised of two pairs of monomers with a greater amount of intersubunit contacts, suggesting a homotetrameric structure of a ‘‘dimer-of-dimers’’. The dimers are held together through an interaction between Arg-438 of one subunit and Glu-433 of the neighboring subunit, forming a salt bridge <ref>PMID: 9927652</ref>. The monomeric structure maize’s C4-PEPC monomer consists of an eight-stranded β barrel and 42 α helices <ref name="kai2003"/><ref name="Izui2004">PMID: 15725057</ref>. The <scene name='57/573979/Cv/3'>active site</scene> of each monomer, bound to PEP and the <scene name='57/573979/Cv/3'>Mn+2 ion</scene> cofactor, is located in the C-terminus region of the β barrel <ref name="matsumura2002">PMID: 12467579</ref>. |
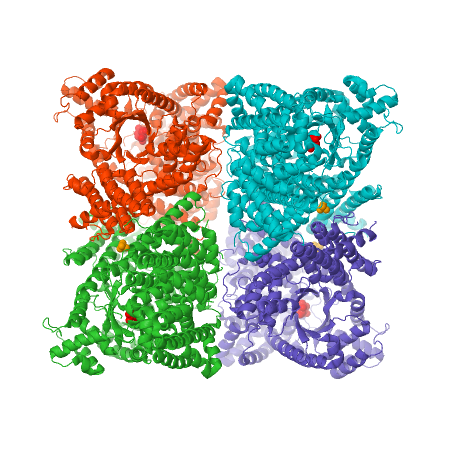
[[Image:4BXC_F.trinervia_PEPC+G6P_crystral_structure.png|center|frame|caption position=bottom|'''Figure 3''' X-ray crystal structure of ''Flaverina trinervia’s'' C4 PEPC bound to glucose 6-phosphate (magenta). Two strongly bound dimers (left and right sides of the structure) form the tetrameric quaternary structure. Adapted from Schlieper et al. 2014. <ref name="schlieper2014">PMID: 24043710</ref>]] | [[Image:4BXC_F.trinervia_PEPC+G6P_crystral_structure.png|center|frame|caption position=bottom|'''Figure 3''' X-ray crystal structure of ''Flaverina trinervia’s'' C4 PEPC bound to glucose 6-phosphate (magenta). Two strongly bound dimers (left and right sides of the structure) form the tetrameric quaternary structure. Adapted from Schlieper et al. 2014. <ref name="schlieper2014">PMID: 24043710</ref>]] | ||
| Line 29: | Line 29: | ||
== Allosteric regulation and reaction mechanism == | == Allosteric regulation and reaction mechanism == | ||
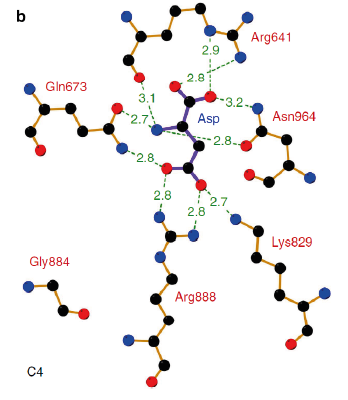
| - | PEPC’s carboxylase activity is regulated by different post-translational mechanisms. In C4 and CAM plants, the phosphorylation of a serine residue near the N terminus (S15 in maize C4-PEPC) activates the enzyme by decreasing its sensitivity to allosteric inhibitors such as aspartate and malate and increasing activation by the positive allosteric regulator glucose 6-phosphate <ref name="O'Leary2011"/>. Studies on'' F. pringlei'' and ''F. trinervia'', have positively identified the residues Arg641, Lys829, Arg888 and Asn964 as binding motif of the negative allosteric inhibitors aspartate and malate <ref name="Paulus2013"/>. Similar studies have also identified the <scene name='57/573979/Cv/ | + | PEPC’s carboxylase activity is regulated by different post-translational mechanisms. In C4 and CAM plants, the phosphorylation of a serine residue near the N terminus (S15 in maize C4-PEPC) activates the enzyme by decreasing its sensitivity to allosteric inhibitors such as aspartate and malate and increasing activation by the positive allosteric regulator glucose 6-phosphate <ref name="O'Leary2011"/>. Studies on'' F. pringlei'' and ''F. trinervia'', have positively identified the residues Arg641, Lys829, Arg888 and Asn964 as binding motif of the negative allosteric inhibitors aspartate and malate <ref name="Paulus2013"/>. Similar studies have also identified the <scene name='57/573979/Cv/8'>aspartate binding site</scene> site in maize <ref name="matsumura2002"/>. In C3 PEPC, Arg884 provides an additional hydrogen bond for inhibitor binding, whereas in C4 PEPC isoforms the substitution of this residue by a glycine, reducing the enzymes sensitivity towards both feedback inhibitors <ref>PMID: 21491491</ref><ref name="Paulus2013"/>. The positive allosteric effector glucose 6-phosphate’s binding site has also been identified in the C4-PEPC of maize. X-ray crystallography of maize’s C4-PEPC in complex with sulfate ion (a positive effector analog of glucose 6-phosphate) revealed that the positive effector was bound to the enzyme at the dimer interface and was surrounded by four positively charged residues (R183, R184, R231, and R372 in the adjacent subunit <ref name="kai2003"/>. |
[[Image:Inhibitor-binding_site_of_Flaverina_trinervia_C4_PEPC.png|center|frame|caption position=bottom|'''Figure 4''' Inhibitor-binding site of ''Flaverina trinervia’s'' C4 PEPC. Adapted from Paulus et al. 2013. <ref name="Paulus2013"/>]] | [[Image:Inhibitor-binding_site_of_Flaverina_trinervia_C4_PEPC.png|center|frame|caption position=bottom|'''Figure 4''' Inhibitor-binding site of ''Flaverina trinervia’s'' C4 PEPC. Adapted from Paulus et al. 2013. <ref name="Paulus2013"/>]] | ||
| Line 60: | Line 60: | ||
'''''Arabidopsis thaliana'''''<br> | '''''Arabidopsis thaliana'''''<br> | ||
| - | [[5fdn]] – AtPEPC 3 + aspartate + citrate<br> | + | [[8oj9]] – AtPEPC 1 <br /> |
| + | [[8ojf]] – AtPEPC 1 + phosphate <br /> | ||
| + | [[8oje]] – AtPEPC 1 + malate <br /> | ||
| + | [[5fdn]] – AtPEPC 3 + aspartate + citrate <br /> | ||
'''''Flaveria pringlei'''''<br> | '''''Flaveria pringlei'''''<br> | ||
Current revision
Contents |
Function
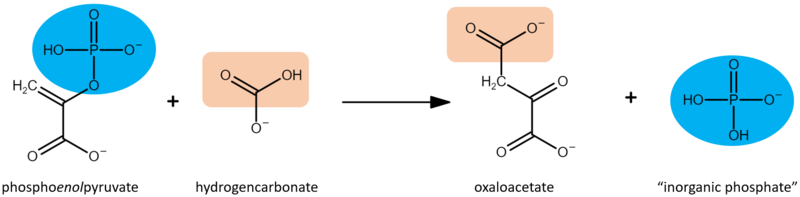
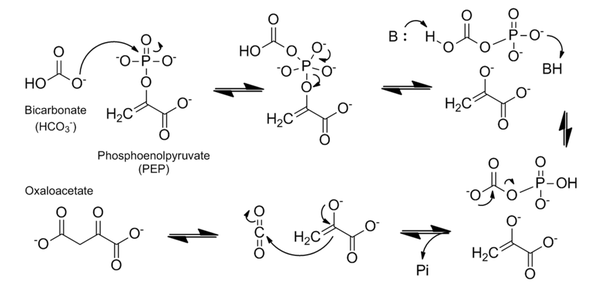
Phosphoenolpyruvate carboxylase (PEPC) catalyzes the irreversible β-carboxylation of PEP in the presence of HCO3- to yield OAA (oxaloacetate) and Pi ( inorganic phosphate), using Mg2 + as a cofactor [1]. It is a key element in C4 carbon fixation [2].
Overview
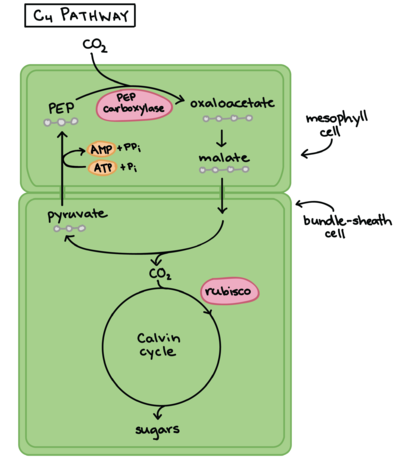
The enzyme phosphoenolpyruvate carboxylase (PEPC) catalyzes the carboxylation of phosphoenolpyruvate to form oxaloacetate, with Mg2+ or Mn2+ as essential cofactors [3][4]. It can be considered the key enzyme in the C4 photosynthesis process, once it’s a central part of the mechanism that makes C4 plants more efficient in carbon fixation compared to classical C3-photosynthetic pathway plants, especially in abiotic stress environments [5]. PEPC is a ubiquitous enzyme, present in the genome of all plants. However, the isoforms found in C4 metabolism plants differ in their kinetic and regulatory characteristics, when compared to C3 orthologs [6]. Among the Flaverina genus of the Asteraceae family, closely related C3 and C4 species are found, providing a good model to study the differences between the two processes. The comparative analysis of Flaverina pringlei (C3) and Flaverina trinervia (C4) PEPC’s, has shown that the exchange of single amino acids can be responsible for the observed differences in saturation kinetics and inhibitor tolerance between PEPC’s of C3 and C4 species [7][6].
PEPC and C4 photosynthesis
In the classical C3 photosynthetic pathway, the RuBisCO enzyme, abundant in leaf mesophyll cells, is responsible for the carboxylation ribulose-1,5-bisphosphate (RuBP) and, therefore, for the assimilation of atmospheric CO2 into 3-phosphoglyceric acid (PGA). However, under high temperature or high concentrations of O2, RuBisCO’s oxygenating activity is favored leading to the oxygenation of RuBP, which produces phosphoglycolate (PG) as well as PGA. While PGA can readily be recycled back to RuBP via the Calvin cycle, PG has to be first metabolized into pyruvate and then to PGA. This process is commonly referred to as photorespiration and it leads to significant losses in freshly assimilated carbon to the atmosphere, essentially diminishing photosynthetic efficiency [8][5].
Plants with C4 photosynthetic metabolism, on the other hand, show very low levels of photorespiration, and consequently are more efficient in terms of carbon fixation [8]. That is made possible by a complex reorganization of leaf anatomy and metabolism in a CO2-concentrating mechanism that inhibits the photorespiratory pathway [5][6]. Without this characteristics, efficient carbon fixation in the mesophyll and subsequent carbon concentration in the BS wouldn’t be possible. Therefore, the evolution of PEPC’s C4 isoforms is one of the most important steps in the establishment of the C4 pathway [2].
| |||||||||||
3D structures of phosphoenolpyruvate carboxylase
Escherichia coli
1jqn – EcPEPC + aspartate + PEP analog
1qb4 – EcPEPC + aspartate
1fiy – EcPEPC + aspartate
Zea mays
1jqo – ZmPEPC
6v3o – ZmPEPC + citrate
5vyj – ZmPEPC + glycine
6u2t – ZmPEPC + malate
6mgi – ZmPEPC + α-D-glucose-6-phosphate
Flaverina trinervia
4bxc – FtPEPC + α-D-glucose-6-phosphate
4bxh – FtPEPC
Arabidopsis thaliana
8oj9 – AtPEPC 1
8ojf – AtPEPC 1 + phosphate
8oje – AtPEPC 1 + malate
5fdn – AtPEPC 3 + aspartate + citrate
Flaveria pringlei
3zge – FpPEPC + aspartate
3zgb – FpPEPC + aspartate
Actinomyces israelii
6k31 – PEPC + CO
Clostridium perfringens
3odm – PEPC + malonate
Updated on 24-August-2023
References
- ↑ 1.0 1.1 1.2 O'Leary B, Park J, Plaxton WC. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J. 2011 May 15;436(1):15-34. PMID:21524275 doi:10.1042/BJ20110078
- ↑ 2.0 2.1 Sage RF. The evolution of C(4) photosynthesis. New Phytol. 2004 Feb;161(2):341-370. PMID:33873498 doi:10.1111/j.1469-8137.2004.00974.x
- ↑ 3.0 3.1 3.2 3.3 3.4 Kai Y, Matsumura H, Izui K. Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Arch Biochem Biophys. 2003 Jun 15;414(2):170-9. PMID:12781768 doi:10.1016/s0003-9861(03)00170-x
- ↑ Svensson P, Bläsing OE, Westhoff P. Evolution of C4 phosphoenolpyruvate carboxylase. Arch Biochem Biophys. 2003 Jun 15;414(2):180-8. PMID:12781769 doi:10.1016/s0003-9861(03)00165-6
- ↑ 5.0 5.1 5.2 Sage RF, Sage TL, Kocacinar F. Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Biol. 2012;63:19-47. PMID:22404472 doi:10.1146/annurev-arplant-042811-105511
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 Paulus JK, Schlieper D, Groth G. Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat Commun. 2013 Feb 26;4:1518. doi: 10.1038/ncomms2504. PMID:23443546 doi:http://dx.doi.org/10.1038/ncomms2504
- ↑ 7.0 7.1 Bläsing OE, Westhoff P, Svensson P. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. J Biol Chem. 2000 Sep 8;275(36):27917-23. PMID:10871630 doi:10.1074/jbc.M909832199
- ↑ 8.0 8.1 Bauwe, H. Chapter 6 Photorespiration: The Bridge to C4 Photosynthesis. in C4 Photosynthesis and Related CO2 Concentrating Mechanisms (eds. Raghavendra, A. S. & Sage, R. F.) 81–108 (Springer Netherlands, 2011). doi:10.1007/978-90-481-9407-0_6.
- ↑ Kai Y, Matsumura H, Inoue T, Terada K, Nagara Y, Yoshinaga T, Kihara A, Tsumura K, Izui K. Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition. Proc Natl Acad Sci U S A. 1999 Feb 2;96(3):823-8. PMID:9927652
- ↑ 10.0 10.1 10.2 10.3 Izui K, Matsumura H, Furumoto T, Kai Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol. 2004;55:69-84. PMID:15725057 doi:10.1146/annurev.arplant.55.031903.141619
- ↑ 11.0 11.1 Matsumura H, Xie Y, Shirakata S, Inoue T, Yoshinaga T, Ueno Y, Izui K, Kai Y. Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. Structure. 2002 Dec;10(12):1721-30. PMID:12467579
- ↑ Schlieper D, Foerster K, Paulus JK, Groth G. Resolving the activation site of positive regulators in plant phosphoenolpyruvate carboxylase. Mol Plant. 2013 Sep 16. PMID:24043710 doi:10.1093/mp/sst130
- ↑ Dharmarajan L, Kraszewski JL, Mukhopadhyay B, Dunten PW. Structure of an archaeal-type phosphoenolpyruvate carboxylase sensitive to inhibition by aspartate. Proteins. 2011 Feb 3. doi: 10.1002/prot.23006. PMID:21491491 doi:10.1002/prot.23006
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Lucas Xavier da Cunha, Michal Harel, Alexander Berchansky, Joel L. Sussman