Phosphoglycerate dehydrogenase
From Proteopedia
(Difference between revisions)
| (18 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | < | + | <StructureSection load='' size='350' side='right' caption='Phosphoglycerate dehydrogenase complex with hydroxypyruvic acid phosphate and tartarate (PDB entry [[3ddn]])' scene='49/491889/Cv/1'> |
| - | + | == Function == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''' | + | |
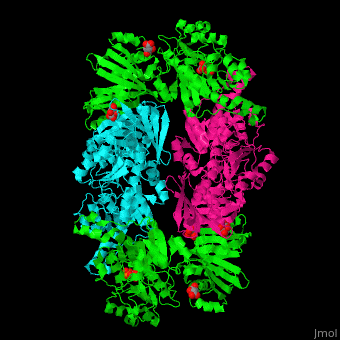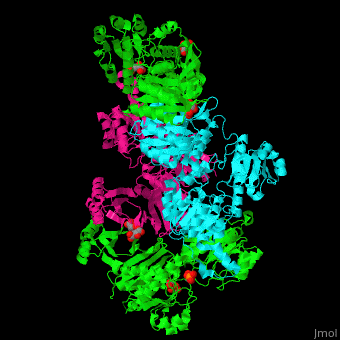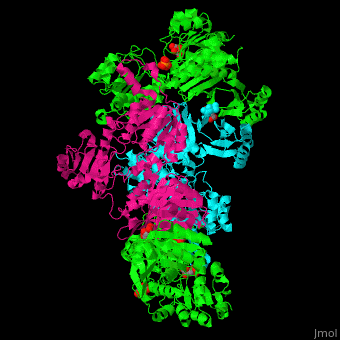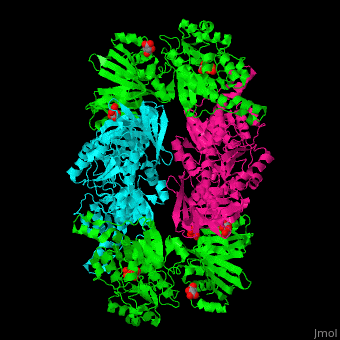
| + | '''Phosphoglycerate dehydrogenase''' (PGDH) catalyzes the conversion of 3-phosphoglycerate into 3-phosphohydroxypyruvate. This is the first step of serine biosynthesis<ref>PMID:11567059</ref>. NAD is used as cofactor. | ||
| + | |||
| + | See also:[[3-Phosphoglycerates: serine, glycine, cysteine]] | ||
| + | |||
| + | == Relevance == | ||
| + | In some cancer cells there is an elevation in PGDH level and hence an increase in serine biosynthesis<ref>PMID:26831078</ref>. | ||
| + | |||
| + | == Structural highlights == | ||
| + | The active site of ''Mycobacterium tuberculosis'' PGDH located in the cleft between the substrate and the NAD-binding domains contains the <scene name='49/491889/Cv/3'>catalytic dyad Glu:His and the substrate</scene><ref>PMID:18627175</ref>. | ||
| + | |||
| + | See [[Rossmann fold]]. | ||
| + | </StructureSection> | ||
==3D structures of phosphoglycerate dehydrogenase== | ==3D structures of phosphoglycerate dehydrogenase== | ||
| - | [[1ygy]], [[3dc2]] – MtPGDH – ''Mycobacterium tuberculosis''<br /> | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} |
| - | [[3evt]] - PGDH – ''Lactobacillus plantarum''<br /> | + | {{#tree:id=OrganizedByTopic|openlevels=0| |
| - | [[3gg9]] - PGDH – ''Ralstonia solanacearum''<br /> | + | |
| - | [[3k5p]] - PGDH (mutant) – ''Brucella melitensis'' | + | *'''Phosphoglycerate dehydrogenase''' |
| + | |||
| + | **[[1ygy]], [[3dc2]] – MtPGDH – ''Mycobacterium tuberculosis''<br /> | ||
| + | **[[3evt]] - PGDH – ''Lactobacillus plantarum''<br /> | ||
| + | **[[3gg9]] - PGDH – ''Ralstonia solanacearum''<br /> | ||
| + | **[[3k5p]] - PGDH (mutant) – ''Brucella melitensis'' | ||
| - | + | *Phosphoglycerate dehydrogenase binary complex | |
| - | [[1sc6]], [[2p9c]], [[2p9e]], [[2p9g]] - EcPGDH (mutant) + NAD – ''Escherichia coli''<br /> | + | **[[2g76]], [[7cvp]] - h3-PGDH + NAD – human<br /> |
| - | [[1yba]] - EcPGDH + NAD<br /> | + | **[[5n53]], [[5nzo]], [[5nzp]], [[5nzq]], [[5ofv]], [[5ofw]], [[7ewh]] – h3-PGDH + drug<br /> |
| - | [[1wwk]] - PGDH + NAD – ''Pyrococcus horikoshii''<br /> | + | **[[5ofm]] – h3-PGDH + amino-methyl-indole<br /> |
| - | + | **[[7va1]] – h3-PGDH + thiazole derivative<br /> | |
| - | [[2ekl]] - PGDH + NAD – ''Sulfolobus tokodaii''<br /> | + | **[[7dkm]] – h3-PGDH + inhibitor<br /> |
| - | [[3ddn]] - MtPGDH + hydroxypyruvic acid phosphate | + | **[[1sc6]], [[2p9c]], [[2p9e]], [[2p9g]] - EcPGDH (mutant) + NAD – ''Escherichia coli''<br /> |
| + | **[[1yba]] - EcPGDH + NAD<br /> | ||
| + | **[[1wwk]] - PGDH + NAD – ''Pyrococcus horikoshii''<br /> | ||
| + | **[[2ekl]] - PGDH + NAD – ''Sulfolobus tokodaii''<br /> | ||
| + | **[[3ddn]] - MtPGDH + hydroxypyruvic acid phosphate | ||
| - | + | *Phosphoglycerate dehydrogenase ternary complex | |
| - | [[1psd]] - EcPGDH + serine + NAD <br /> | + | **[[5n6c]] – h3-PGDH + NAD + tartrate<br /> |
| - | [[2pa3]] - EcPGDH (mutant) + serine + NAD | + | **[[6cwa]] – h3-PGDH + NAD + 3PG<br /> |
| + | **[[6plf]], [[6plg]] – h3-PGDH + indole amide derivative<br /> | ||
| + | **[[6rih]], [[6rj2]], [[6rj3]], [[6rj5]], [[6rj6]] – h3-PGDH + inhibitor<br /> | ||
| + | **[[1psd]] - EcPGDH + serine + NAD <br /> | ||
| + | **[[2pa3]] - EcPGDH (mutant) + serine + NAD<br /> | ||
| + | }} | ||
| + | == References == | ||
| + | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of phosphoglycerate dehydrogenase
Updated on 27-August-2023
References
- ↑ Yamasaki M, Yamada K, Furuya S, Mitoma J, Hirabayashi Y, Watanabe M. 3-Phosphoglycerate dehydrogenase, a key enzyme for l-serine biosynthesis, is preferentially expressed in the radial glia/astrocyte lineage and olfactory ensheathing glia in the mouse brain. J Neurosci. 2001 Oct 1;21(19):7691-704. PMID:11567059
- ↑ Mullarky E, Lucki NC, Beheshti Zavareh R, Anglin JL, Gomes AP, Nicolay BN, Wong JC, Christen S, Takahashi H, Singh PK, Blenis J, Warren JD, Fendt SM, Asara JM, DeNicola GM, Lyssiotis CA, Lairson LL, Cantley LC. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci U S A. 2016 Feb 16;113(7):1778-83. doi:, 10.1073/pnas.1521548113. Epub 2016 Feb 1. PMID:26831078 doi:http://dx.doi.org/10.1073/pnas.1521548113
- ↑ Dey S, Burton RL, Grant GA, Sacchettini JC. Structural analysis of substrate and effector binding in Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase. Biochemistry. 2008 Aug 12;47(32):8271-82. Epub 2008 Jul 16. PMID:18627175 doi:10.1021/bi800212b