Phosphoserine aminotransferase
From Proteopedia
(Difference between revisions)
| (20 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
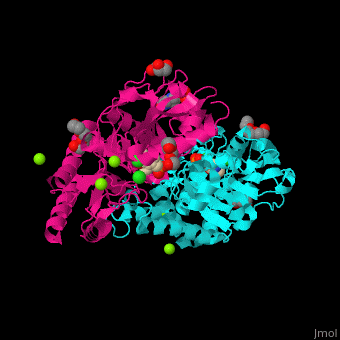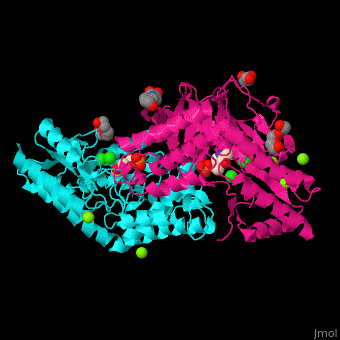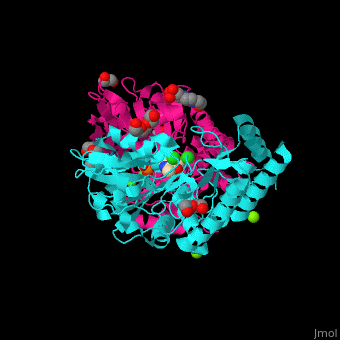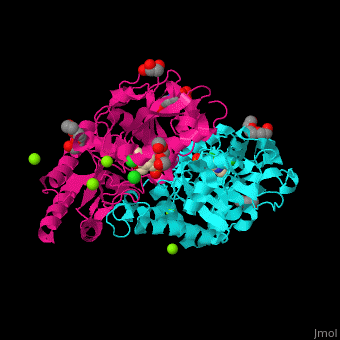
| - | + | <StructureSection load='' size='350' side='right' caption='Phosphoserine aminotransferase dimer complex with pyridoxal phosphate, triethylene glycol, Hepes, Cl- and Mg+2 ions (PDB entry [[1w23]])' scene='48/488518/Cv/1'> | |
| + | == Function == | ||
| + | '''Phosphoserine aminotransferase''' (PSAT) catalyzes the reversible conversion of phosphoserine and oxoglutarate to produce glutamate and 3-phosphonooxypyruvate. PSAT is part of the phosphoserine biosynthesis<ref>PMID:10637769</ref>. Pyridoxal phosphate (vitamin B6, PLP) is a cofactor of PAT. | ||
| - | + | == Relevance == | |
| - | + | PSAT is over-expressed in colon tumors and increases their chemoresistance. PSAT1 inhibition is being tested as drug treatment<ref>PMID:18221502</ref>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == Structural highlights == | ||
| + | PSAT overall structure shows a <scene name='48/488518/Cv/8'>small domain and a large one</scene>. The <scene name='48/488518/Cv/9'>active site</scene> contains the <scene name='48/488518/Cv/10'>PLP co-factor bound to lysine side chain</scene><ref>PMID:15608117</ref>. Water molecules are shown as red spheres. | ||
| + | |||
| + | </StructureSection> | ||
==3D structures of phosphoserine aminotransferase== | ==3D structures of phosphoserine aminotransferase== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | *'''Phosphoserine aminotransferase''' | ||
| - | [[3m5u]] – PSAT – ''Campylobacter jejuni''<br /> | + | **[[8i28]] - yPSAT – yeast<br /> |
| - | [[3qm2]] – PSAT – ''Salmonella enterica'' | + | **[[3m5u]] – PSAT – ''Campylobacter jejuni''<br /> |
| + | **[[3qm2]] – PSAT – ''Salmonella enterica''<br /> | ||
| + | **[[5yii]] - EhPSAT – ''Entamoeba histolytica''<br /> | ||
| - | + | *Phosphoserine aminotransferase complex with pyridoxal phosphate | |
| - | [[1bt4]], [[1w3u]], [[2c0r]] – PSAT (mutant) + PLP – ''Bacillus circulans''<br /> | + | **[[3e77]] – hPSAT + PLP – human<br /> |
| - | [[1bjn]] – EcPSAT + PLP – ''Escherichia coli''<br /> | + | **[[8a5v]] – hPSAT1 + PLP derivative <br /> |
| - | [[1w23]], [[2bhx]], [[2bi1]], [[2bi2]], [[2bi3]], [[2bi5]], [[2bi9]], [[2bia]], [[2bie]], [[2big]] - PSAT + PLP – ''Bacillus alcalophilus''<br /> | + | **[[1bt4]], [[1w3u]], [[2c0r]] – PSAT (mutant) + PLP – ''Bacillus circulans''<br /> |
| - | [[ | + | **[[1bjn]] – EcPSAT + PLP – ''Escherichia coli''<br /> |
| - | [[ | + | **[[1w23]], [[2bhx]], [[2bi1]], [[2bi2]], [[2bi3]], [[2bi5]], [[2bi9]], [[2bia]], [[2bie]], [[2big]], [[4azj]], [[4azk]] - PSAT + PLP – ''Bacillus alcalophilus''<br /> |
| - | [[ | + | **[[3ffr]] – PSAT + PLP – ''Cytophaga hutchinsonii''<br /> |
| + | **[[3qbo]] – PSAT + PLP <br /> | ||
| + | **[[2fyf]] - PSAT + PLP – ''Mycobacterium tuberculosis''<br /> | ||
| + | **[[6czz]] – AtPSAT1 + PLP – ''Arabidopsis thaliana''<br /> | ||
| + | **[[6czx]], [[6czy]] – AtPSAT1 + PLP derivative <br /> | ||
| + | **[[5yb0]] - EhPSAT + PLP <br /> | ||
| + | **[[5yd2]] - EhPSAT (mutant) + PLP<br /> | ||
| + | **[[6xdk]] - PSAT + PLP – ''Stenotrophomonas maltophilia''<br /> | ||
| + | **[[5f8v]] - PSAT + PLP – ''Trichomonas vaginalis<''br /> | ||
| + | **[[7t7j]] - PSAT + PLP – ''Klebsiella pneuoniae<''br /> | ||
| - | + | *Phosphoserine aminotransferase ternary complex | |
| - | [[1bjo]] - EcPSAT + α-methyl-glutamate + PLP | + | **[[8a5w]] - hPSAT + PLP derivative + phosphoserine <br /> |
| + | **[[1bjo]] - EcPSAT + α-methyl-glutamate + PLP<br /> | ||
| + | **[[4xk1]] - PSAT + PLP + glutamate – ''Pseudomonas aeruginosa''<br /> | ||
| + | }} | ||
| + | == References == | ||
| + | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of phosphoserine aminotransferase
Updated on 29-August-2023
References
- ↑ Basurko MJ, Marche M, Darriet M, Cassaigne A. Phosphoserine aminotransferase, the second step-catalyzing enzyme for serine biosynthesis. IUBMB Life. 1999 Nov;48(5):525-9. PMID:10637769 doi:http://dx.doi.org/10.1080/713803557
- ↑ Vie N, Copois V, Bascoul-Mollevi C, Denis V, Bec N, Robert B, Fraslon C, Conseiller E, Molina F, Larroque C, Martineau P, Del Rio M, Gongora C. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. 2008 Jan 25;7:14. doi: 10.1186/1476-4598-7-14. PMID:18221502 doi:http://dx.doi.org/10.1186/1476-4598-7-14
- ↑ Dubnovitsky AP, Kapetaniou EG, Papageorgiou AC. Enzyme adaptation to alkaline pH: atomic resolution (1.08 A) structure of phosphoserine aminotransferase from Bacillus alcalophilus. Protein Sci. 2005 Jan;14(1):97-110. PMID:15608117 doi:14/1/97