|
|
| Line 3: |
Line 3: |
| | <StructureSection load='3ciy' size='340' side='right'caption='[[3ciy]], [[Resolution|resolution]] 3.41Å' scene=''> | | <StructureSection load='3ciy' size='340' side='right'caption='[[3ciy]], [[Resolution|resolution]] 3.41Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[3ciy]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Lk3_transgenic_mice Lk3 transgenic mice]. The November 2011 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Toll-like Receptors'' by David Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2011_11 10.2210/rcsb_pdb/mom_2011_11]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3CIY OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3CIY FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[3ciy]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Mus_musculus Mus musculus]. The November 2011 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Toll-like Receptors'' by David Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2011_11 10.2210/rcsb_pdb/mom_2011_11]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3CIY OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3CIY FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BMA:BETA-D-MANNOSE'>BMA</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=FUC:ALPHA-L-FUCOSE'>FUC</scene>, <scene name='pdbligand=FUL:BETA-L-FUCOSE'>FUL</scene>, <scene name='pdbligand=MAN:ALPHA-D-MANNOSE'>MAN</scene>, <scene name='pdbligand=NDG:2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE'>NDG</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.41Å</td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat"><div style='overflow: auto; max-height: 3em;'>[[3cig|3cig]], [[2a0z|2a0z]]</div></td></tr>
| + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BMA:BETA-D-MANNOSE'>BMA</scene>, <scene name='pdbligand=FUC:ALPHA-L-FUCOSE'>FUC</scene>, <scene name='pdbligand=FUL:BETA-L-FUCOSE'>FUL</scene>, <scene name='pdbligand=MAN:ALPHA-D-MANNOSE'>MAN</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=NDG:2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE'>NDG</scene></td></tr> |
| - | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">Tlr3 ([https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=10090 LK3 transgenic mice])</td></tr>
| + | |
| | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3ciy FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3ciy OCA], [https://pdbe.org/3ciy PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3ciy RCSB], [https://www.ebi.ac.uk/pdbsum/3ciy PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3ciy ProSAT]</span></td></tr> | | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3ciy FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3ciy OCA], [https://pdbe.org/3ciy PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3ciy RCSB], [https://www.ebi.ac.uk/pdbsum/3ciy PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3ciy ProSAT]</span></td></tr> |
| | </table> | | </table> |
| | == Function == | | == Function == |
| - | [[https://www.uniprot.org/uniprot/TLR3_MOUSE TLR3_MOUSE]] Key component of innate and adaptive immunity. TLRs (Toll-like receptors) control host immune response against pathogens through recognition of molecular patterns specific to microorganisms. TLR3 is a nucleotide-sensing TLR which is activated by double-stranded RNA, a sign of viral infection. Acts via MYD88 and TRAF6, leading to NF-kappa-B activation, cytokine secretion and the inflammatory response (By similarity).<ref>PMID:14993594</ref>
| + | [https://www.uniprot.org/uniprot/TLR3_MOUSE TLR3_MOUSE] Key component of innate and adaptive immunity. TLRs (Toll-like receptors) control host immune response against pathogens through recognition of molecular patterns specific to microorganisms. TLR3 is a nucleotide-sensing TLR which is activated by double-stranded RNA, a sign of viral infection. Acts via MYD88 and TRAF6, leading to NF-kappa-B activation, cytokine secretion and the inflammatory response (By similarity).<ref>PMID:14993594</ref> |
| | == Evolutionary Conservation == | | == Evolutionary Conservation == |
| | [[Image:Consurf_key_small.gif|200px|right]] | | [[Image:Consurf_key_small.gif|200px|right]] |
| Line 40: |
Line 39: |
| | </StructureSection> | | </StructureSection> |
| | [[Category: Large Structures]] | | [[Category: Large Structures]] |
| - | [[Category: Lk3 transgenic mice]] | + | [[Category: Mus musculus]] |
| | [[Category: RCSB PDB Molecule of the Month]] | | [[Category: RCSB PDB Molecule of the Month]] |
| | [[Category: Toll-like Receptors]] | | [[Category: Toll-like Receptors]] |
| - | [[Category: Botos, I]] | + | [[Category: Botos I]] |
| - | [[Category: Davies, D R]] | + | [[Category: Davies DR]] |
| - | [[Category: Leonard, J N]] | + | [[Category: Leonard JN]] |
| - | [[Category: Liu, L]] | + | [[Category: Liu L]] |
| - | [[Category: Segal, D M]] | + | [[Category: Segal DM]] |
| - | [[Category: Shiloach, J]] | + | [[Category: Shiloach J]] |
| - | [[Category: Wang, Y]] | + | [[Category: Wang Y]] |
| - | [[Category: Dsrna]]
| + | |
| - | [[Category: Glycoprotein]]
| + | |
| - | [[Category: Immune response]]
| + | |
| - | [[Category: Immune system-rna complex]]
| + | |
| - | [[Category: Inflammatory response]]
| + | |
| - | [[Category: Innate immunity]]
| + | |
| - | [[Category: Leucine rich repeat]]
| + | |
| - | [[Category: Leucine-rich repeat]]
| + | |
| - | [[Category: Lrr]]
| + | |
| - | [[Category: Membrane]]
| + | |
| - | [[Category: Protein-dsrna complex]]
| + | |
| - | [[Category: Receptor]]
| + | |
| - | [[Category: Tlr]]
| + | |
| - | [[Category: Transmembrane]]
| + | |
| Structural highlights
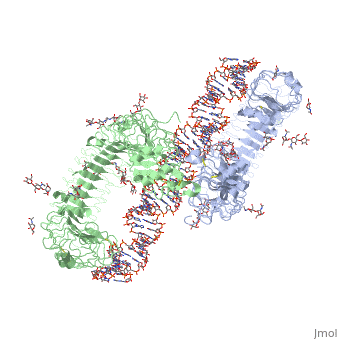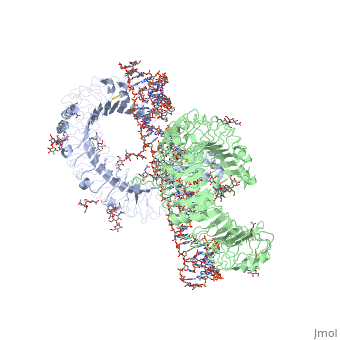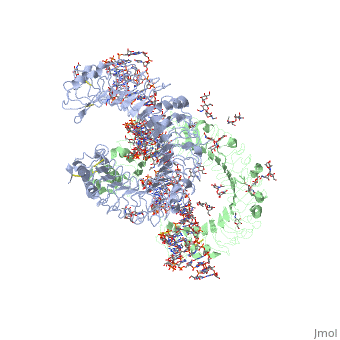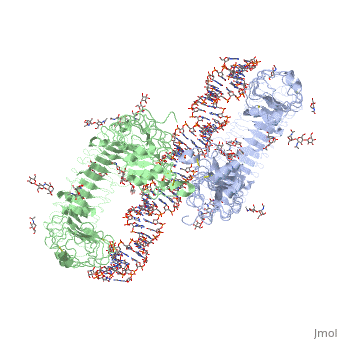
3ciy is a 4 chain structure with sequence from Mus musculus. The November 2011 RCSB PDB Molecule of the Month feature on Toll-like Receptors by David Goodsell is 10.2210/rcsb_pdb/mom_2011_11. Full crystallographic information is available from OCA. For a guided tour on the structure components use FirstGlance.
| | Method: | X-ray diffraction, Resolution 3.41Å |
| Ligands: | , , , , , |
| Resources: | FirstGlance, OCA, PDBe, RCSB, PDBsum, ProSAT |
Function
TLR3_MOUSE Key component of innate and adaptive immunity. TLRs (Toll-like receptors) control host immune response against pathogens through recognition of molecular patterns specific to microorganisms. TLR3 is a nucleotide-sensing TLR which is activated by double-stranded RNA, a sign of viral infection. Acts via MYD88 and TRAF6, leading to NF-kappa-B activation, cytokine secretion and the inflammatory response (By similarity).[1]
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
Toll-like receptor 3 (TLR3) recognizes double-stranded RNA (dsRNA), a molecular signature of most viruses, and triggers inflammatory responses that prevent viral spread. TLR3 ectodomains (ECDs) dimerize on oligonucleotides of at least 40 to 50 base pairs in length, the minimal length required for signal transduction. To establish the molecular basis for ligand binding and signaling, we determined the crystal structure of a complex between two mouse TLR3-ECDs and dsRNA at 3.4 angstrom resolution. Each TLR3-ECD binds dsRNA at two sites located at opposite ends of the TLR3 horseshoe, and an intermolecular contact between the two TLR3-ECD C-terminal domains coordinates and stabilizes the dimer. This juxtaposition could mediate downstream signaling by dimerizing the cytoplasmic Toll interleukin-1 receptor (TIR) domains. The overall shape of the TLR3-ECD does not change upon binding to dsRNA.
Structural basis of toll-like receptor 3 signaling with double-stranded RNA.,Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR Science. 2008 Apr 18;320(5874):379-81. PMID:18420935[2]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, Shamel L, Sovath S, Goode J, Alexopoulou L, Flavell RA, Beutler B. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci U S A. 2004 Mar 9;101(10):3516-21. Epub 2004 Mar 1. PMID:14993594 doi:http://dx.doi.org/10.1073/pnas.0400525101
- ↑ Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008 Apr 18;320(5874):379-81. PMID:18420935 doi:320/5874/379
|