Poly(A) RNA polymerase
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
'''Poly(A) RNA polymerase protein Cid1''' (Caffeine-Induced Death protein 1) is involved in cell cycle arrest where it inhibits unscheduled mitosis<ref>PMID:17072891</ref>. Cid1 is found in fission yeast. Cid1 exists in 3 forms: '''apo I''' consists of 4 subunits; '''apo II''' consists of 2 subunits; '''UTP-bound''' form consists of 4 subunits. <br /> | '''Poly(A) RNA polymerase protein Cid1''' (Caffeine-Induced Death protein 1) is involved in cell cycle arrest where it inhibits unscheduled mitosis<ref>PMID:17072891</ref>. Cid1 is found in fission yeast. Cid1 exists in 3 forms: '''apo I''' consists of 4 subunits; '''apo II''' consists of 2 subunits; '''UTP-bound''' form consists of 4 subunits. <br /> | ||
| - | '''Poly(A) RNA polymerase Gld2''' is responsible for poly(A) tail lengthening. Gld2 activity in the brain is required for long-term memory<ref>PMID: | + | '''Poly(A) RNA polymerase Gld2''' is responsible for poly(A) tail lengthening. Gld2 activity in the brain is required for long-term memory<ref>PMID:15987818</ref>. |
== Structural highlights == | == Structural highlights == | ||
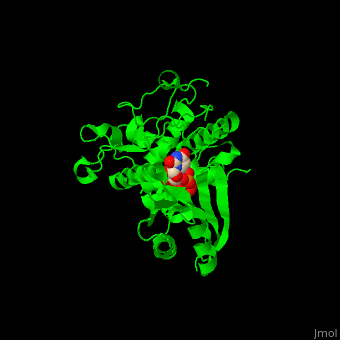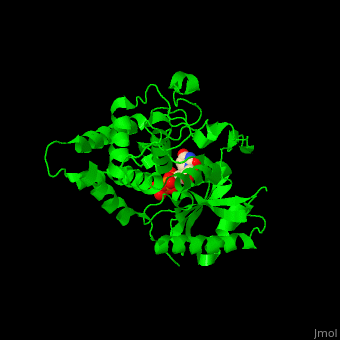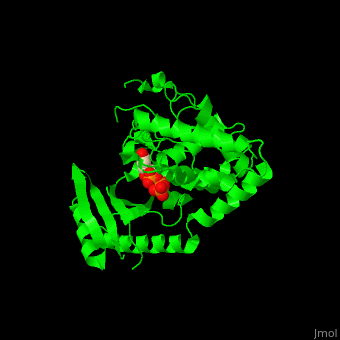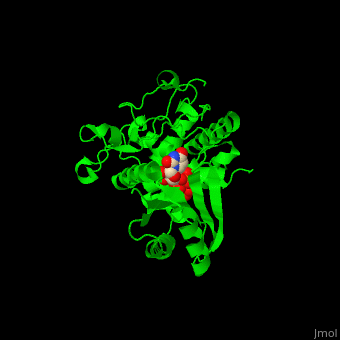
| - | The UTP binding site of Cid1 is between the N-terminal and C-terminal. The <scene name='70/706259/Cv/ | + | The UTP binding site of Cid1 is between the N-terminal and C-terminal. The <scene name='70/706259/Cv/5'>active site</scene> contains the <scene name='70/706259/Cv/6'>catalytic triad of aspartic acids</scene> (in magenta) in the N-terminal and the <scene name='70/706259/Cv/7'>His residue</scene> (in cyan) whose two conformations select between binding of UTP and ATP to Cid1 <ref>PMID:22751018</ref>. Water molecules are shown as red spheres. |
</StructureSection> | </StructureSection> | ||
| - | == 3D Structures of poly(A) RNA polymerase | + | == 3D Structures of poly(A) RNA polymerase == |
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| Line 29: | Line 29: | ||
**[[4nku]] – fyCid1 residues 40-377 (mutant) + dinucleotide<br /> | **[[4nku]] – fyCid1 residues 40-377 (mutant) + dinucleotide<br /> | ||
| - | *Poly(A) RNA polymerase Gld2 | + | *Poly(A) RNA polymerase Gld2 or terminal nucleotidyltransferase 2 |
| + | **[[6lbj]] - Gld2 residues 143-484 (mutant) – mouse<br /> | ||
| + | **[[6lbk]] - Gld2 residues 131-484 (mutant) – rat<br /> | ||
**[[5jnb]] - CeGld2 residues 546-923 + RNP-8 – ''Caenorhabditis elegans''<br /> | **[[5jnb]] - CeGld2 residues 546-923 + RNP-8 – ''Caenorhabditis elegans''<br /> | ||
**[[4zrl]] – CeGld2 residues 527-923 + defective in germ line development protein<br /> | **[[4zrl]] – CeGld2 residues 527-923 + defective in germ line development protein<br /> | ||
| + | |||
| + | *Poly(A) RNA polymerase | ||
| + | |||
| + | **[[3pq1]] – hPAP (mutant) - human<br /> | ||
}} | }} | ||
Current revision
| |||||||||||
3D Structures of poly(A) RNA polymerase
Updated on 03-September-2023
References
- ↑ Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006 Oct 15;23(13):991-1000. PMID:17072891 doi:http://dx.doi.org/10.1002/yea.1408
- ↑ Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005 Jul;11(7):1117-30. PMID:15987818 doi:http://dx.doi.org/11/7/1117
- ↑ Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol. 2012 Jul 1. doi: 10.1038/nsmb.2329. PMID:22751018 doi:10.1038/nsmb.2329
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky