We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3igc
From Proteopedia
(Difference between revisions)
(New page: '''Unreleased structure''' The entry 3igc is ON HOLD Authors: Perry, K., Hwang, Y., Bushman, F.D., Van Duyne, G.D. Description: Smallpox virus topoisomerase-DNA transition state ''Pag...) |
|||
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | '''Unreleased structure''' | ||
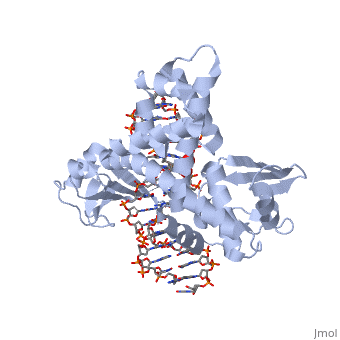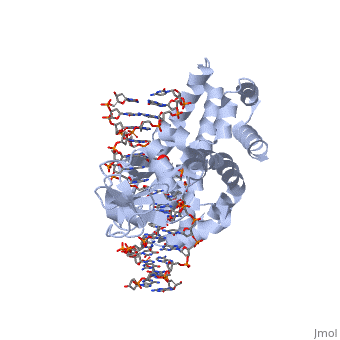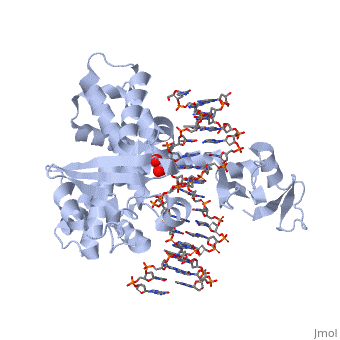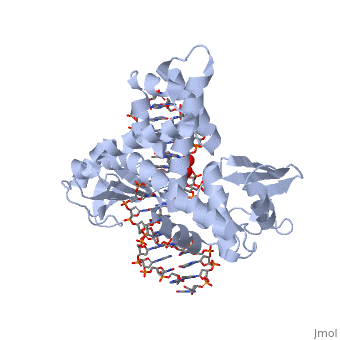
| - | + | ==Smallpox virus topoisomerase-DNA transition state== | |
| + | <StructureSection load='3igc' size='340' side='right'caption='[[3igc]], [[Resolution|resolution]] 2.10Å' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[3igc]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Variola_virus Variola virus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3IGC OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3IGC FirstGlance]. <br> | ||
| + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.1Å</td></tr> | ||
| + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=VO4:VANADATE+ION'>VO4</scene></td></tr> | ||
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3igc FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3igc OCA], [https://pdbe.org/3igc PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3igc RCSB], [https://www.ebi.ac.uk/pdbsum/3igc PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3igc ProSAT]</span></td></tr> | ||
| + | </table> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/TOP1_VAR67 TOP1_VAR67] Releases the supercoiling and torsional tension of DNA introduced during the DNA replication and transcription by transiently cleaving and rejoining one strand of the DNA duplex. Introduces a single-strand break via transesterification at the specific target site 5'-[CT]CCTTp site in duplex DNA. The scissile phosphodiester is attacked by the catalytic tyrosine of the enzyme, resulting in the formation of a DNA-(3'-phosphotyrosyl)-enzyme intermediate and the expulsion of a 5'-OH DNA strand. The free DNA strand then undergoes passage around the unbroken strand thus removing DNA supercoils. Finally, in the religation step, the DNA 5'-OH attacks the covalent intermediate to expel the active-site tyrosine and restore the DNA phosphodiester backbone (By similarity). | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ig/3igc_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3igc ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Poxviruses encode their own type IB topoisomerases (TopIBs), which release superhelical tension generated by replication and transcription of their genomes. To investigate the reaction catalyzed by viral TopIBs, we have determined the structure of a variola virus topoisomerase-DNA complex trapped as a vanadate transition state mimic. The structure reveals how the viral TopIB enzymes are likely to position the DNA duplex for ligation following relaxation of supercoils and identifies the sources of friction observed in single-molecule experiments that argue against free rotation. The structure also identifies a conformational change in the leaving group sugar that must occur prior to cleavage and reveals a mechanism for promoting ligation following relaxation of supercoils that involves an Asp-minor groove interaction. Overall, the new structural data support a common catalytic mechanism for the TopIB superfamily but indicate distinct methods for controlling duplex rotation in the small versus large enzyme subfamilies. | ||
| - | + | Insights from the structure of a smallpox virus topoisomerase-DNA transition state mimic.,Perry K, Hwang Y, Bushman FD, Van Duyne GD Structure. 2010 Jan 13;18(1):127-37. PMID:20152159<ref>PMID:20152159</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| + | </div> | ||
| + | <div class="pdbe-citations 3igc" style="background-color:#fffaf0;"></div> | ||
| - | + | ==See Also== | |
| + | *[[Topoisomerase 3D structures|Topoisomerase 3D structures]] | ||
| + | == References == | ||
| + | <references/> | ||
| + | __TOC__ | ||
| + | </StructureSection> | ||
| + | [[Category: Large Structures]] | ||
| + | [[Category: Variola virus]] | ||
| + | [[Category: Bushman FD]] | ||
| + | [[Category: Hwang Y]] | ||
| + | [[Category: Perry K]] | ||
| + | [[Category: Van Duyne GD]] | ||
Current revision
Smallpox virus topoisomerase-DNA transition state
| |||||||||||