TAL effector
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 8: | Line 8: | ||
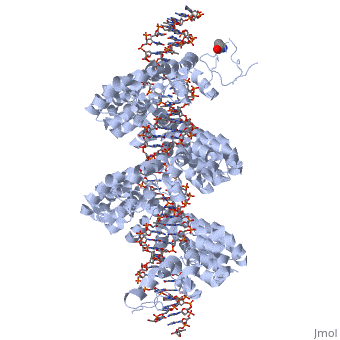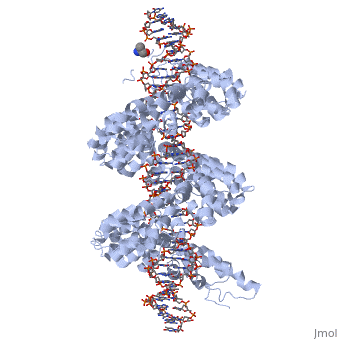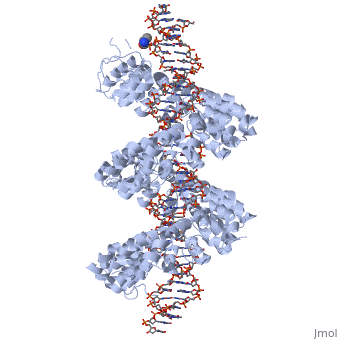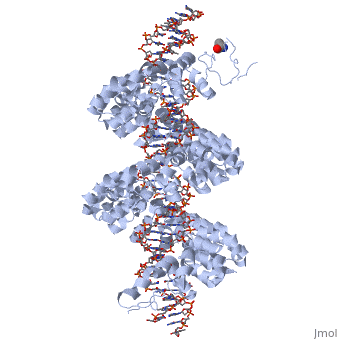
<br>[[Image:overview.jpg|left|300px|thumb|Fig.1 Structure of the PthXo1 DNA binding region in complex with its target site. The coloring of individual repeats matches the schematic in XXX]]<br> | <br>[[Image:overview.jpg|left|300px|thumb|Fig.1 Structure of the PthXo1 DNA binding region in complex with its target site. The coloring of individual repeats matches the schematic in XXX]]<br> | ||
| - | + | {{Clear}} | |
==DNA binding module with marked RVD== | ==DNA binding module with marked RVD== | ||
<br>[[Image:sanjana.png|left|400px|thumb|Fig.2]]<br> | <br>[[Image:sanjana.png|left|400px|thumb|Fig.2]]<br> | ||
| + | {{Clear}} | ||
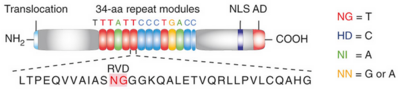
'''Figure 2''': Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = <scene name='Sandbox_Reserved_701/T_blau/1'>T</scene>, HD = <scene name='Sandbox_Reserved_701/C_schwarz/1'>C</scene>, NI = <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>, and NN = <scene name='Sandbox_Reserved_701/G_gelb/1'>G</scene> or <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>. The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site.<ref> PMID:022223736 </ref> | '''Figure 2''': Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = <scene name='Sandbox_Reserved_701/T_blau/1'>T</scene>, HD = <scene name='Sandbox_Reserved_701/C_schwarz/1'>C</scene>, NI = <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>, and NN = <scene name='Sandbox_Reserved_701/G_gelb/1'>G</scene> or <scene name='Sandbox_Reserved_701/A_gruen/1'>A</scene>. The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site.<ref> PMID:022223736 </ref> | ||
| Line 16: | Line 17: | ||
<br>[[Image:Mak_An.png|right|400px|thumb|Fig.3]]<br> | <br>[[Image:Mak_An.png|right|400px|thumb|Fig.3]]<br> | ||
| + | {{Clear}} | ||
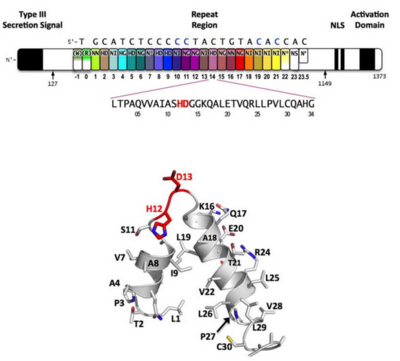
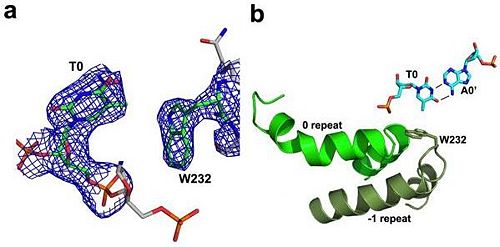
'''Figure 3''': Domain organization of PthXo1 and structure of a single TAL effector repeat | '''Figure 3''': Domain organization of PthXo1 and structure of a single TAL effector repeat | ||
TAL effectors contain N-terminal signals for bacterial type III secretion, tandem repeats that specify the target nucleotide sequence, nuclear localization signals, and a C-terminal region that is required for transcriptional activation. PthXo1 contains 23.5 canonical repeats (color coded to match Figure 3) that contact the DNA target found in the promoter of the rice Os8N3 gene (17). Blue bases correspond to positions in the target where the match between protein and DNA differs from the optimal match specified by the recognition code (3,4). Arrows indicate the start and end of the crystallized protein construct. In the structure, repeats 22 to 23.5 are poorly ordered, as are the C-termini of the two N-terminal cryptic repeats. The sequence and structure of a representative repeat (#14) is shown; RVD residues (HD) that recognize cytosine are red.<ref> PMID:22222791 </ref> | TAL effectors contain N-terminal signals for bacterial type III secretion, tandem repeats that specify the target nucleotide sequence, nuclear localization signals, and a C-terminal region that is required for transcriptional activation. PthXo1 contains 23.5 canonical repeats (color coded to match Figure 3) that contact the DNA target found in the promoter of the rice Os8N3 gene (17). Blue bases correspond to positions in the target where the match between protein and DNA differs from the optimal match specified by the recognition code (3,4). Arrows indicate the start and end of the crystallized protein construct. In the structure, repeats 22 to 23.5 are poorly ordered, as are the C-termini of the two N-terminal cryptic repeats. The sequence and structure of a representative repeat (#14) is shown; RVD residues (HD) that recognize cytosine are red.<ref> PMID:22222791 </ref> | ||
<br>[[Image:nihms396484f3.jpg|left|400px|thumb|Fig.4]]<br> | <br>[[Image:nihms396484f3.jpg|left|400px|thumb|Fig.4]]<br> | ||
| + | {{Clear}} | ||
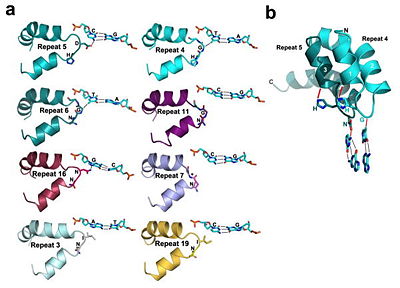
'''Figure 4''': All of the repeats in the DNA-bound PthXo1 structure form highly similar two-helix bundles (Figure 1c). The helices span positions 3 to 11 and 14 to 33, locating the RVD in a loop between them. A proline located at position 27, creates a kink in the second helix that appears to be critical for the sequential packing and association of tandem repeats with the DNA double helix. The packing of consecutive helices within and between individual repeats is left-handed, in contrast to the right-handed packing of helices found in TPR proteins (10). The modular architecture of the TAL effector repeats is reminiscent of the mitochondrial transcription terminator [http://www.proteopedia.org/wiki/index.php/3n6s mTERF] (11) and the [http://www.proteopedia.org/wiki/index.php/TRAP RNA-binding attenuation protein TRAP] (12). | '''Figure 4''': All of the repeats in the DNA-bound PthXo1 structure form highly similar two-helix bundles (Figure 1c). The helices span positions 3 to 11 and 14 to 33, locating the RVD in a loop between them. A proline located at position 27, creates a kink in the second helix that appears to be critical for the sequential packing and association of tandem repeats with the DNA double helix. The packing of consecutive helices within and between individual repeats is left-handed, in contrast to the right-handed packing of helices found in TPR proteins (10). The modular architecture of the TAL effector repeats is reminiscent of the mitochondrial transcription terminator [http://www.proteopedia.org/wiki/index.php/3n6s mTERF] (11) and the [http://www.proteopedia.org/wiki/index.php/TRAP RNA-binding attenuation protein TRAP] (12). | ||
| Line 50: | Line 53: | ||
==3D structures of TAL effectors== | ==3D structures of TAL effectors== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
[[3ugm]] - XoTAL pthxo1 + DNA - ''Xanthomonas oryzae''<br /> | [[3ugm]] - XoTAL pthxo1 + DNA - ''Xanthomonas oryzae''<br /> | ||
| Line 56: | Line 60: | ||
[[3v6t]], [[4osh]], [[4osi]], [[4osj]], [[4osk]], [[4osl]] – XoTAL dHax3 + DNA <br /> | [[3v6t]], [[4osh]], [[4osi]], [[4osj]], [[4osk]], [[4osl]] – XoTAL dHax3 + DNA <br /> | ||
[[4gjr]], [[4gjp]], [[4osm]], [[4osq]], [[4osr]], [[4oss]], [[4ost]], [[4osv]], [[4osw]], [[4osz]], [[4ot0]], [[4ot3]], [[4oto]] – XoTAL dHax3 (mutant) + DNA<br /> | [[4gjr]], [[4gjp]], [[4osm]], [[4osq]], [[4osr]], [[4oss]], [[4ost]], [[4osv]], [[4osw]], [[4osz]], [[4ot0]], [[4ot3]], [[4oto]] – XoTAL dHax3 (mutant) + DNA<br /> | ||
| + | [[6jtq]], [[6jw0]], [[6jvz]], [[6jw4]], [[6jw3]], [[6jw2]], [[6jw5]], [[6jw1]] – XoTAL + DNA <br /> | ||
| + | [[6lew]] - TAL + DNA - ''Xanthomonas campestris''<br /> | ||
==Reference== | ==Reference== | ||
Current revision
| |||||||||||
3D structures of TAL effectors
Updated on 12-December-2023
3ugm - XoTAL pthxo1 + DNA - Xanthomonas oryzae
3v6p - XoTAL dHax3
4gg4 – XoTAL dHax3 + DNA + RNA
3v6t, 4osh, 4osi, 4osj, 4osk, 4osl – XoTAL dHax3 + DNA
4gjr, 4gjp, 4osm, 4osq, 4osr, 4oss, 4ost, 4osv, 4osw, 4osz, 4ot0, 4ot3, 4oto – XoTAL dHax3 (mutant) + DNA
6jtq, 6jw0, 6jvz, 6jw4, 6jw3, 6jw2, 6jw5, 6jw1 – XoTAL + DNA
6lew - TAL + DNA - Xanthomonas campestris
Reference
- Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The Crystal Structure of TAL Effector PthXo1 Bound to Its DNA Target. Science. 2012 Jan 5. PMID:22223736 doi:10.1126/science.1216211
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012 Jan 5;7(1):171-92. doi: 10.1038/nprot.2011.431. PMID:22222791 doi:10.1038/nprot.2011.431
- Barberousse V, Sacco D, Dellacherie E. Chromatographic evaluation of the binding of haemoglobin to polyanionic polymers. J Chromatogr. 1986 Nov 7;369(1):244-7. PMID:2432079
Contributors
Fabian Stritt, Philipp Warmer
Proteopedia Page Contributors and Editors (what is this?)
Philipp Warmer, Michal Harel, Jaime Prilusky, OCA, Alexander Berchansky