Tumor susceptibility gene 101
From Proteopedia
(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
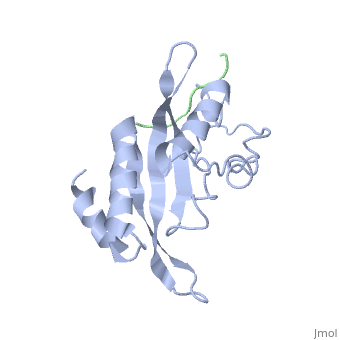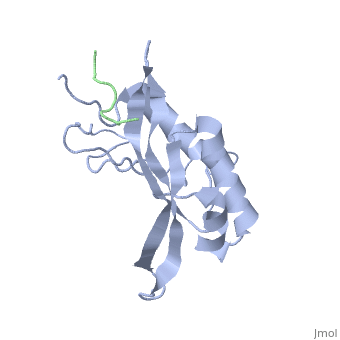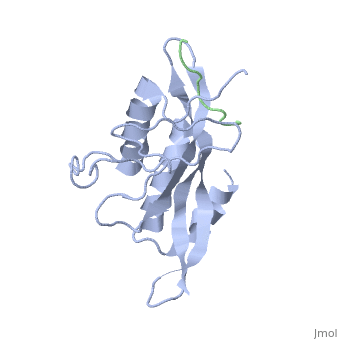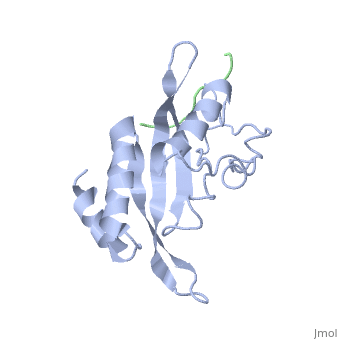
| - | + | <StructureSection load='1m4p' size='340' side='right' caption='Human tumor susceptibility gene 101 protein (grey) complex with GAG polyprotein peptide (green) (PDB code [[1mp4]])' scene=''> | |
| - | <StructureSection load='1m4p' size='340' side='right' caption='Human tumor susceptibility gene 101 protein (grey) complex with GAG polyprotein peptide (PDB code [[1mp4]])' scene=''> | + | The product of the human '''tumor susceptibility gene 101''' (Tsg101) is a component of the endocytic sorting complex required for transport (ESCRT-1 complex), which recognizes ubiquitinated protein cargo and facilitates cargo delivery to sub-cellular compartments for degradation. Ironically, Tsg101 also is required for release of the human immunodeficiency virus (HIV) from cells. |
| - | The product of the human tumor susceptibility gene 101 (Tsg101) is a component of the endocytic sorting complex required for transport (ESCRT-1 complex), which recognizes ubiquitinated protein cargo and facilitates cargo delivery to sub-cellular compartments for degradation. Ironically, Tsg101 also is required for release of the human immunodeficiency virus (HIV) from cells. | + | |
| - | + | [[Image:HIV for Proteopedia.JPG|left|300 px]] | |
| + | {{Clear}} | ||
__TOC__ | __TOC__ | ||
==Background and History== | ==Background and History== | ||
| Line 33: | Line 34: | ||
Polymers of [http://www.medicaldictionaryweb.com/N-substituted+Glycines-definition/ N-substituted glycine] (NSG) residues have recently emerged as an important class of peptide inhibitors that effectively can substitute for proline in WW and SH3 domain-binding proteins. In the Tsg101 UEV, the Ala 9 - Pro 10 binding pocket resembles the x-proline pockets of WW and SH3 domains. SH3 domains are about 50 to 70 amino acids long and are often present in proteins responsible for eukaryotic signal transduction and are found in the cytoskeleton. These domains recognize proline rich ligands with a PxxP motif (where x is any amino acid and P is proline). WW domains are the smallest protein-binding modules and are made up of approximately 40 amino acids. The name refers to two tryptophan residues spaced approximately 20 to 22 amino acids apart, which also recognize proline-rich ligands. While WW domains resemble SH3 domains functionally by binding to proline rich ligands, WW domains bind to PPxY motifs (where Y is tyrosine). The key proline residues in ligands that bind to WW and SH3 domains take on [http://en.wikipedia.org/wiki/Polyproline_helix polyproline helical] conformations like PTAP and bind with the proline ring wedged between two perpendicularly situated aromatic rings on the domains. The positions of the key prolines and the two aromatic rings are conserved across the Tsg101 UEV P(S/T)AP, WW PPxY and SH3 PxxP complexes. This is significant because in order for viral budding to occur, Tsg101 has to bind to the PTAP region of the Gag and N-substituted 'peptoid' inhibitors have been identified to successfully compete with the binding of proline rich ligands to WW and SH3 domains. The similarities between Tsg101 and SH3 domains in their recognition of key proline residues suggest that the replacement of proline residues with NSG constructs may eventually lead to the preparation of PTAP-based Tsg101 binding inhibitors that can potentially prevent viral budding. | Polymers of [http://www.medicaldictionaryweb.com/N-substituted+Glycines-definition/ N-substituted glycine] (NSG) residues have recently emerged as an important class of peptide inhibitors that effectively can substitute for proline in WW and SH3 domain-binding proteins. In the Tsg101 UEV, the Ala 9 - Pro 10 binding pocket resembles the x-proline pockets of WW and SH3 domains. SH3 domains are about 50 to 70 amino acids long and are often present in proteins responsible for eukaryotic signal transduction and are found in the cytoskeleton. These domains recognize proline rich ligands with a PxxP motif (where x is any amino acid and P is proline). WW domains are the smallest protein-binding modules and are made up of approximately 40 amino acids. The name refers to two tryptophan residues spaced approximately 20 to 22 amino acids apart, which also recognize proline-rich ligands. While WW domains resemble SH3 domains functionally by binding to proline rich ligands, WW domains bind to PPxY motifs (where Y is tyrosine). The key proline residues in ligands that bind to WW and SH3 domains take on [http://en.wikipedia.org/wiki/Polyproline_helix polyproline helical] conformations like PTAP and bind with the proline ring wedged between two perpendicularly situated aromatic rings on the domains. The positions of the key prolines and the two aromatic rings are conserved across the Tsg101 UEV P(S/T)AP, WW PPxY and SH3 PxxP complexes. This is significant because in order for viral budding to occur, Tsg101 has to bind to the PTAP region of the Gag and N-substituted 'peptoid' inhibitors have been identified to successfully compete with the binding of proline rich ligands to WW and SH3 domains. The similarities between Tsg101 and SH3 domains in their recognition of key proline residues suggest that the replacement of proline residues with NSG constructs may eventually lead to the preparation of PTAP-based Tsg101 binding inhibitors that can potentially prevent viral budding. | ||
| - | + | </StructureSection> | |
==3D structures of tumor susceptibility gene 101== | ==3D structures of tumor susceptibility gene 101== | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| Line 40: | Line 41: | ||
*Tumor susceptibility gene 101 | *Tumor susceptibility gene 101 | ||
| - | **[[1kpp]], [[1kpq]], [[2f0r]] – hTsg101 UEV domain - human<br /> | + | **[[1kpp]], [[1kpq]], [[2f0r]], [[4yc1]], [[7zlx]] – hTsg101 UEV domain residues 1-145 - human<br /> |
**[[3obs]] – hTsg101 UEV domain (mutant) <br /> | **[[3obs]] – hTsg101 UEV domain (mutant) <br /> | ||
| + | **[[5vkg]] – hTsg101 UEV domain (mutant) + tenatoprazole <br /> | ||
**[[3iv1]] – hTsg101 coiled-coil domain <br /> | **[[3iv1]] – hTsg101 coiled-coil domain <br /> | ||
| Line 47: | Line 49: | ||
**[[4eje]] – hTsg101 UEV domain + ebola peptide <br /> | **[[4eje]] – hTsg101 UEV domain + ebola peptide <br /> | ||
| - | **[[1m4p]], [[1m4q]] – hTsg101 UEV domain + gag protein peptide <br /> | + | **[[1m4p]], [[1m4q]], [[4zny]] – hTsg101 UEV domain + gag protein peptide <br /> |
**[[3obu]], [[3obx]], [[3p9g]], [[3p9h]] – hTsg101 UEV domain (mutant) + gag protein peptide <br /> | **[[3obu]], [[3obx]], [[3p9g]], [[3p9h]] – hTsg101 UEV domain (mutant) + gag protein peptide <br /> | ||
| - | **[[1s1q]] – hTsg101 UEV domain + ubiquitin <br /> | + | **[[7nlc]] – hTsg101 UEV domain + orthohepevirus peptide <br /> |
| + | **[[1s1q]], [[6ud0]] – hTsg101 UEV domain + ubiquitin <br /> | ||
**[[3obq]] – hTsg101 UEV domain (mutant) + HRS PSAP peptide <br /> | **[[3obq]] – hTsg101 UEV domain (mutant) + HRS PSAP peptide <br /> | ||
| + | **[[6vme]] – hTsg101 residues 308-388 in ESCRT-I headpiece<br /> | ||
}} | }} | ||
| Line 74: | Line 78: | ||
* VerPlank L, Bouamr F, LaGrassa T, Agresta B, Kikonyogo A, Leis J, Carter C. 2001. ''Tsg101, a homolgue of ubiquitin-conjugating (E2) enzymes, binds the L domain of HIV type 1 Pr55Gag.'' Proceeding of the National Academy of Science 98:7724-7729. | * VerPlank L, Bouamr F, LaGrassa T, Agresta B, Kikonyogo A, Leis J, Carter C. 2001. ''Tsg101, a homolgue of ubiquitin-conjugating (E2) enzymes, binds the L domain of HIV type 1 Pr55Gag.'' Proceeding of the National Academy of Science 98:7724-7729. | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of tumor susceptibility gene 101
Updated on 06-February-2024
Additional Resources
For additional information, see: Human Immunodeficiency Virus
References
- Freed E. 2003. The HIV-Tsg101 Interface: recent advances in a budding field. Trends in Microbiology 11:56-59.
- Liu F, Stephen Ag, Adamson C, Gousset K, Aman MJ, Freed EO, Fisher RJ, Burke TR, Jr. 2005. Hydrazone and Hydrazide-Containing N-Substituted Glycines as Peptoid Surrogates for Expedited Library Synthesis: Application to the Preparation of Tsg101-Directed HIV-1 Budding Antagonist. Org Lett 8:5163-5168.
- Macias M, Wiesnera S, Sudolb M. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. Protein Domains 513:30-37.
- Pornillos O, Alam S, Davis D, Sundquist W. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nature Structural Biology 9:812–817.
- Pornillos O, Higginson D, Stray K, Fisher R, Garrus J, Payne M, He G, Wang H, Morham S, Sundquist W. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. The Journal of Cell Biology 162:425-434.
- Sudol M, Recinos C, Abraczinskas J, Humbert J, Farooq A. 2005. WW or WoW: The domains in a Union of Bliss. Life 57:773-778.
- Tavassoli A, Lu Q, Gam J, Pan H, Benkovic S, Cohen S. 2008. Inhibition of HIV Budding by a Genetically Selected Cyclic Peptide Targeting the Gag-TSG101 Interaction. American Chemical Society,Chemical Biology 3:757-764.
- VerPlank L, Bouamr F, LaGrassa T, Agresta B, Kikonyogo A, Leis J, Carter C. 2001. Tsg101, a homolgue of ubiquitin-conjugating (E2) enzymes, binds the L domain of HIV type 1 Pr55Gag. Proceeding of the National Academy of Science 98:7724-7729.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Gisselle Medina, Eran Hodis, Lois A. Fridmann, Nargiza Mahmudova, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky, David Canner, Motunrayo Mobolaji-Lawal, Melanie Ng