UDP-3-O-acyl-N-acetylglucosamine deacetylase
From Proteopedia
(Difference between revisions)
| (10 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
== Function == | == Function == | ||
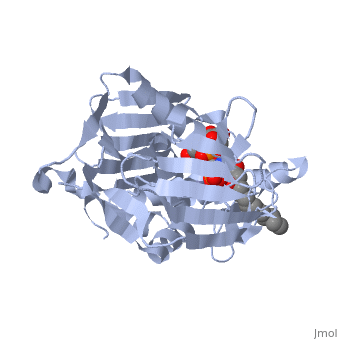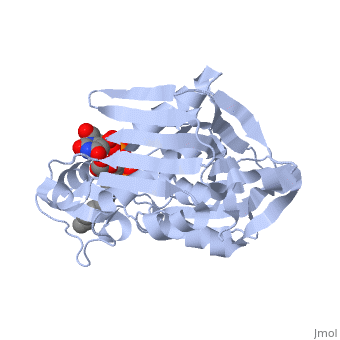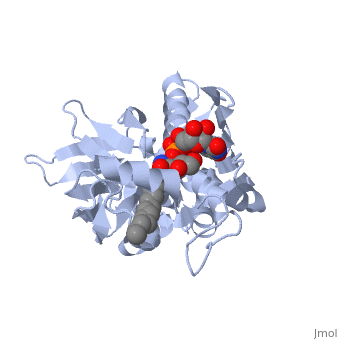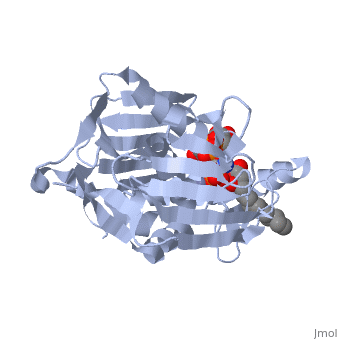
| - | '''UDP-3-O-acyl-N-acetylglucosamine deacetylase''' (LpxC) is a Zn-dependent protein which participates in the biosynthesis of lipid A. Lipid A anchors lipopolysaccharides into the membrane of Gram negative bacteria. LpxC catalyzes the conversion of UDP-3-O-[(3R)-3-hydroxymyristoyl]-N-acetylglucosamine to UDP-3-O-[(3R)-3-hydroxymyristoyl]-N-glucosamine. | + | '''UDP-3-O-acyl-N-acetylglucosamine deacetylase''' (LpxC) is a Zn-dependent protein which participates in the biosynthesis of lipid A. Lipid A anchors lipopolysaccharides into the membrane of Gram negative bacteria. LpxC catalyzes the conversion of UDP-3-O-[(3R)-3-hydroxymyristoyl]-N-acetylglucosamine to UDP-3-O-[(3R)-3-hydroxymyristoyl]-N-glucosamine<ref>PMID:18289052</ref>. |
| + | <scene name='59/595824/Cv/1'>Structure of E. coli UDP-3-O-acyl-N-acetylglucosamine deacetylase complex with Zn+2 ion and UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine</scene> (PDB code [[4mdt]]). | ||
| + | *<scene name='59/595824/Cv/2'>Zn coordination site</scene>. | ||
| + | *<scene name='59/595824/Cv/3'>PO4 interactions</scene>. Water molecules are shown as red spheres. | ||
| + | *<scene name='59/595824/Cv/6'>UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine binding site</scene>. | ||
| + | *<scene name='59/595824/Cv/7'>Whole binding site</scene>. | ||
| + | *<scene name='59/595824/Cv/8'>Surface representation</scene>. | ||
| + | *<scene name='59/595824/Cv/9'>UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine is located in long tunnel</scene>. | ||
=== Para-(benzoyl)-phenylalanine as a potential inhibitor against LpxC of ''Leptospira spp.'': Homology modeling, docking and molecular dynamics study <ref>doi 10.1080/07391102.2012.758056</ref>=== | === Para-(benzoyl)-phenylalanine as a potential inhibitor against LpxC of ''Leptospira spp.'': Homology modeling, docking and molecular dynamics study <ref>doi 10.1080/07391102.2012.758056</ref>=== | ||
''Leptospira interrogans'', a Gram-negative bacterial pathogen is the main cause of human leptospirosis. Lipid A is a highly immunoreactive endotoxic center of lipopolysaccharide (LPS) that anchors LPS into the outer membrane of Leptospira. Discovery of compounds inhibiting lipid-A biosynthetic pathway would be promising for dissolving the structural integrity of membrane leading to cell lysis and death of Leptospira. <scene name='Journal:JBSD:10/Cv/2'>LpxC</scene>, a unique enzyme of lipid-A biosynthetic pathway was identified as common drug target of ''Leptospira''. Herein, homology modeling, docking, and molecular dynamics (MD) simulations were employed to discover potential inhibitors of LpxC. A <scene name='Journal:JBSD:10/Cv/3'>reliable tertiary structure of LpxC in complex with inhibitor BB-78485</scene> was constructed in Modeller 9v8. <scene name='Journal:JBSD:10/Cv/5'>Click here to see animated comparison</scene> between active site of target and template (LpxC from ''Pseudomonas aeruginosa'', PDB entry [[2ves]]) in the presence of BB-78485. The color code representations as <span style="color:deeppink;background-color:black;font-weight:bold;">deep pink: target</span>; <span style="color:lime;background-color:black;font-weight:bold;">green: template</span>; <span style="color:cyan;background-color:black;font-weight:bold;">cyan: BB-78485 in complex with target</span>; and <font color='darkmagenta'><b>darkmagenta: BB-78485 in complex with template</b></font>. Zn ion represented as sphere. A data-set of BB-78485 structural analogs were docked with LpxC in Maestro v9.2 virtual screening workflow, which implements three stage Glide docking protocol. Twelve lead molecules with better XP Gscore compared to BB-78485 were proposed as potential inhibitors of LpxC. Para-(benzoyl)-phenylalanine – that showed lowest XP Gscore (−10.35 kcal/mol) – was predicted to have best binding affinity towards LpxC. MD simulations were performed for LpxC and para-(benzoyl)-phenylalanine docking complex in Desmond v3.0. Trajectory analysis showed the docking complex and inter-molecular interactions was stable throughout the entire production part of MD simulations. The results indicate para-(benzoyl)-phenylalanine as a potent drug molecule against leptospirosis. | ''Leptospira interrogans'', a Gram-negative bacterial pathogen is the main cause of human leptospirosis. Lipid A is a highly immunoreactive endotoxic center of lipopolysaccharide (LPS) that anchors LPS into the outer membrane of Leptospira. Discovery of compounds inhibiting lipid-A biosynthetic pathway would be promising for dissolving the structural integrity of membrane leading to cell lysis and death of Leptospira. <scene name='Journal:JBSD:10/Cv/2'>LpxC</scene>, a unique enzyme of lipid-A biosynthetic pathway was identified as common drug target of ''Leptospira''. Herein, homology modeling, docking, and molecular dynamics (MD) simulations were employed to discover potential inhibitors of LpxC. A <scene name='Journal:JBSD:10/Cv/3'>reliable tertiary structure of LpxC in complex with inhibitor BB-78485</scene> was constructed in Modeller 9v8. <scene name='Journal:JBSD:10/Cv/5'>Click here to see animated comparison</scene> between active site of target and template (LpxC from ''Pseudomonas aeruginosa'', PDB entry [[2ves]]) in the presence of BB-78485. The color code representations as <span style="color:deeppink;background-color:black;font-weight:bold;">deep pink: target</span>; <span style="color:lime;background-color:black;font-weight:bold;">green: template</span>; <span style="color:cyan;background-color:black;font-weight:bold;">cyan: BB-78485 in complex with target</span>; and <font color='darkmagenta'><b>darkmagenta: BB-78485 in complex with template</b></font>. Zn ion represented as sphere. A data-set of BB-78485 structural analogs were docked with LpxC in Maestro v9.2 virtual screening workflow, which implements three stage Glide docking protocol. Twelve lead molecules with better XP Gscore compared to BB-78485 were proposed as potential inhibitors of LpxC. Para-(benzoyl)-phenylalanine – that showed lowest XP Gscore (−10.35 kcal/mol) – was predicted to have best binding affinity towards LpxC. MD simulations were performed for LpxC and para-(benzoyl)-phenylalanine docking complex in Desmond v3.0. Trajectory analysis showed the docking complex and inter-molecular interactions was stable throughout the entire production part of MD simulations. The results indicate para-(benzoyl)-phenylalanine as a potent drug molecule against leptospirosis. | ||
| - | </StructureSection> | ||
== 3D Structures of UDP-3-O-acyl-N-acetylglucosamine deacetylase == | == 3D Structures of UDP-3-O-acyl-N-acetylglucosamine deacetylase == | ||
| + | |||
| + | [[UDP-3-O-acyl-N-acetylglucosamine deacetylase 3D structures]] | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| Line 29: | Line 37: | ||
**[[1xxe]] – AaLpxC + Zn + inhibitor - NMR <br /> | **[[1xxe]] – AaLpxC + Zn + inhibitor - NMR <br /> | ||
| - | **[[3p76]], [[5dro]], [[5drp]], [[5drq]], [[5drr]], [[4u3b]], [[4u3d]] – AaLpxC + Zn + inhibitor <br /> | + | **[[3p76]], [[5dro]], [[5drp]], [[5drq]], [[5drr]], [[4u3b]], [[4u3d]], [[6ih0]] – AaLpxC + Zn + inhibitor <br /> |
| - | **[[2go4]], [[3p3c]] – AaLpxC (mutant) + Zn + inhibitor <br /> | + | **[[2go4]], [[3p3c]], [[5u86]] – AaLpxC (mutant) + Zn + inhibitor <br /> |
**[[2go3]] – AaLpxC (mutant) + Zn + myristate + cacodylate + imidazole<br /> | **[[2go3]] – AaLpxC (mutant) + Zn + myristate + cacodylate + imidazole<br /> | ||
**[[2o3z]] – AaLpxC (mutant) + Zn + benzoate derivative<br /> | **[[2o3z]] – AaLpxC (mutant) + Zn + benzoate derivative<br /> | ||
**[[2jt2]] – AaLpxC (mutant) + Zn + antibiotic - NMR<br /> | **[[2jt2]] – AaLpxC (mutant) + Zn + antibiotic - NMR<br /> | ||
**[[4oze]] – AaLpxC + Zn + native product<br /> | **[[4oze]] – AaLpxC + Zn + native product<br /> | ||
| - | **[[ | + | **[[4j3d]], [[4fw3]], [[4fw4]], [[4fw5]], [[4fw6]], [[4fw7]], [[5u3b]], [[5u39]], [[5upg]], [[5vwm]], [[6mae]], [[6cax]], [[6c9c]], [[6dui]], [[6e54]], [[6mo4]], [[6mo5]], [[6mod]], [[6moo]], [[6i46]], [[6u47]], [[6i48]], [[6i49]], [[6i4a]], [[7cic]], [[7ci4]], [[7ci5]], [[7ci6]], [[7ci7]], [[7ci8]], [[7ci9]], [[7cia]], [[7cib]], [[7cid]], [[7cie]], [[7k99]], [[7k9a]] – PaLpxC + Zn + inhibitor – ''Pseudomonas aeruginosa''<br /> |
| - | + | **[[2ves]] – PaLpxC (mutant) + Zn + inhibitor - NMR<br /> | |
| - | **[[ | + | **[[3p3e]], [[3u1y]], [[3uhm]], [[4lcf]], [[4lcg]], [[4lch]], [[4okg]], [[5n8c]] – PaLpxC (mutant) + Zn + inhibitor <br /> |
**[[3nzk]] – LpxC + Zn + inhibitor – ''Yersinia enterocolitica''<br /> | **[[3nzk]] – LpxC + Zn + inhibitor – ''Yersinia enterocolitica''<br /> | ||
**[[3p3g]], [[3ps1]], [[3ps2]], [[3ps3]], [[4mqy]], [[4is9]], [[4isa]] – EcLpxC + Zn + inhibitor <br /> | **[[3p3g]], [[3ps1]], [[3ps2]], [[3ps3]], [[4mqy]], [[4is9]], [[4isa]] – EcLpxC + Zn + inhibitor <br /> | ||
| Line 44: | Line 52: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | + | </StructureSection> | |
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||