UDP-N-acetylglucosamine acyltransferase
From Proteopedia
(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
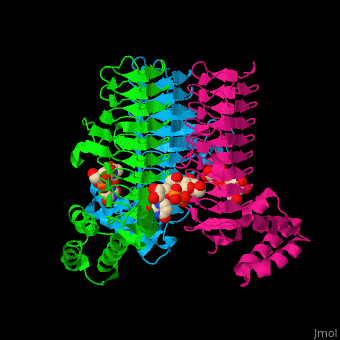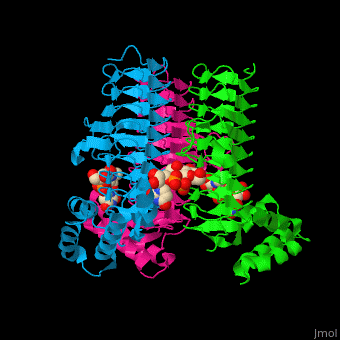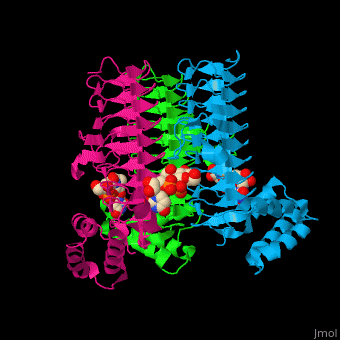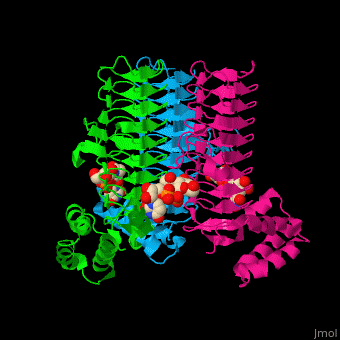
| - | <StructureSection load='' size=' | + | <StructureSection load='' size='350' side='right' caption='E. coli UDP-N-acetylglucosamine acyltransferase complex with UDP-Glc-NAC (PDB code [[2jf3]])' scene='74/749395/Cv/1'> |
== Function == | == Function == | ||
| - | '''UDP-N-acetylglucosamine acyltransferase''' (LpxA) initiates lipid A biosynthesis. It catalyzes the transfer of 3-hydroxymyristic acid from acyl carrier protein to the 3'-hydroxyl group of UDP-GlcNAc<ref>PMID:10480918</ref>. | + | '''UDP-N-acetylglucosamine acyltransferase''' or |
| + | '''Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase''' (LpxA) initiates lipid A biosynthesis. It catalyzes the transfer of 3-hydroxymyristic acid from acyl carrier protein to the 3'-hydroxyl group of UDP-GlcNAc<ref>PMID:10480918</ref>. | ||
== Structural highlights == | == Structural highlights == | ||
| - | The <scene name='74/749395/Cv/ | + | The <scene name='74/749395/Cv/6'>substrate-binding site</scene> of LpxA interacts with the substrate UDP moiety and the Glc-NAc moiety. It contains a <scene name='74/749395/Cv/7'>His-Asp catalytic dyad</scene> where the His acts as a general base and Asp helps to orient the His for participation in the catalysis<ref>PMID:17434525</ref>. |
| - | </StructureSection> | ||
==3D structures of UDP-N-acetylglucosamine acyltransferase== | ==3D structures of UDP-N-acetylglucosamine acyltransferase== | ||
| + | [[UDP-N-acetylglucosamine acyltransferase 3D structures]] | ||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | *UDP-N-acetylglucosamine acyltransferase | ||
| - | |||
| - | **[[5f42]] – LpxA – ''Francicella novicida''<br /> | ||
| - | **[[5dem]] – PaLpxA – ''Pseudomonas aeruginosa''<br /> | ||
| - | **[[4r36]] – BfLpxA – ''Bacterioides fragilis''<br /> | ||
| - | **[[4e6t]], [[4e6u]] – LpxA – ''Acinetobacter baumannii''<br /> | ||
| - | **[[3t57]] – LpxA – ''Arabidopsis thaliana''<br /> | ||
| - | **[[4eqy]] – LpxA – ''Burkholderia thailandensis''<br /> | ||
| - | **[[3r0s]] – LpxA – ''Campylobacter jejuni''<br /> | ||
| - | **[[3hsq]] – LiLpxA – ''Leptospira interrogans''<br /> | ||
| - | **[[2jf2]] – EcLpxA – ''Escherichia coli''<br /> | ||
| - | **[[1lxa]] – EcLpxA (mutant)<br /> | ||
| - | **[[1j2z]] – LpxA – ''Helicobacter pylori''<br /> | ||
| - | **[[5jxx]] – LpxA – ''Moraxella catarrhalis''<br /> | ||
| - | |||
| - | *UDP-N-acetylglucosamine acyltransferase complex | ||
| - | |||
| - | **[[5dep]], [[5dg3]] – PaLpxA + UDP-GlcNAc derivative<br /> | ||
| - | **[[4r37]] – BfLpxA + UDP-GlcNAc <br /> | ||
| - | **[[4j09]] – EcLpxA + RJPXD33 peptide <br /> | ||
| - | **[[2aq9]] – EcLpxA + peptide inhibitor<br /> | ||
| - | **[[2jf3]] – EcLpxA + UDP-GlcNAc <br /> | ||
| - | **[[3i3a]], [[3i3x]] – LiLpxA + sugar nucleotide<br /> | ||
| - | **[[2qia]], [[2qiv]], [[2jf3]] – EcLpxA + sugar nucleotide<br /> | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | </StructureSection> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||