Alcohol dehydrogenase
From Proteopedia
(Difference between revisions)
| (25 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='1hdz' size=' | + | <StructureSection load='1hdz' size='350' side='right' scene='' caption='Human alcohol dehydrogenase dimer with NAD, Zn+2 (grey) and Cl- (green) ions (PDB code [[1hdz]])'> |
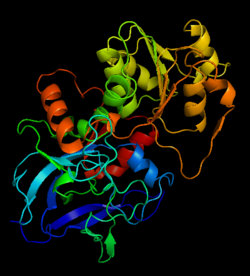
[[Image:1htb2.png|thumb|left|250px|Structure of Alcohol Dehydrogenase]] | [[Image:1htb2.png|thumb|left|250px|Structure of Alcohol Dehydrogenase]] | ||
{{Clear}} | {{Clear}} | ||
| - | '''Alcohol dehydrogenase''' (ADH, EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.1.1.1 1.1.1.1]) is an 80kDa enzyme that catalyzes the 4th step in the metabolism of fructose before [[glycolysis]]. In the 4th step, glyceraldehyde is converted to the glycolytic intermediate DHAP by the NADH-dependent, ADH catalyzed reduction to glycerol.<ref>Voet, et. al. ''Fundamentals of Biochemistry: 3rd Edition''. Hoboken: Wiley & Sons, Inc, 2008.</ref> ADH catalyzes the oxidation of primary and secondary alcohols to their corresponding aldehydes and ketones through a mechanism that involves the removal of a hydrogen | + | __TOC__ |
| + | ==Function== | ||
| + | |||
| + | '''Alcohol dehydrogenase''' (ADH, EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.1.1.1 1.1.1.1]) is an 80kDa enzyme that catalyzes the 4th step in the metabolism of fructose before [[glycolysis]]. In the 4th step, glyceraldehyde is converted to the glycolytic intermediate DHAP by the NADH-dependent, ADH catalyzed reduction to glycerol.<ref>Voet, et. al. ''Fundamentals of Biochemistry: 3rd Edition''. Hoboken: Wiley & Sons, Inc, 2008.</ref> ADH catalyzes the oxidation of primary and secondary alcohols to their corresponding aldehydes and ketones through a mechanism that involves the removal of a hydrogen. More detailed discussions in<br /> | ||
| Line 9: | Line 12: | ||
*[[Alcohol dehydrogenase from Clostridium beijerinckii]]<br /> | *[[Alcohol dehydrogenase from Clostridium beijerinckii]]<br /> | ||
*[[Alcohol dehydrogenase from Entamoeba histolytica]]<br /> | *[[Alcohol dehydrogenase from Entamoeba histolytica]]<br /> | ||
| + | *[[ALDH2]]<br /> | ||
*[[D275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolytica]]<br /> | *[[D275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolytica]]<br /> | ||
*[[Horse Liver Alcohol Dehydrogenase]]<br /> | *[[Horse Liver Alcohol Dehydrogenase]]<br /> | ||
*[[Contribution of Pro275 to the Thermostability of the Alcohol Dehydrogenases]]. | *[[Contribution of Pro275 to the Thermostability of the Alcohol Dehydrogenases]]. | ||
'''Hydroxyacyl-CoA dehydrogenase''' (HADH) catalyzes the conversion of 3-hydroxyacyl-CoA to 3-oxoacyl-CoA. NAD is the cofactor of HADH activity. HADH oxidates straight-chain 3-hydroxyacyl-CoAs in the β-oxidation pathway of fatty acid metabolism. HADH is classified according to its substrate ads short chain (SHCDH) and long chain HADH. HADH deficiency is a genetic disorder. | '''Hydroxyacyl-CoA dehydrogenase''' (HADH) catalyzes the conversion of 3-hydroxyacyl-CoA to 3-oxoacyl-CoA. NAD is the cofactor of HADH activity. HADH oxidates straight-chain 3-hydroxyacyl-CoAs in the β-oxidation pathway of fatty acid metabolism. HADH is classified according to its substrate ads short chain (SHCDH) and long chain HADH. HADH deficiency is a genetic disorder. | ||
| + | |||
| + | '''alcohol dehydrogenase I, II, III, IV''' differ by their tissue specificity. | ||
| + | |||
| + | '''Secondary alcohol dehydrogenase''' catalyses the reduction of acetone to isopropanol<ref>PMID: 22686835</ref>. | ||
| + | |||
| + | '''NADP-dependent alcohol dehydrogenase''' catalyses the redox equilibria between aldehydes or ketones and the corresponding primary or secondary alcohols <ref>PMID: 8349550</ref>. | ||
| + | |||
| + | '''Quinone-dependent alcohol dehydrogenase''' oxidizes primary, secondary alcohols, aldehydes, polysaccharides and cyclodextrins <ref>PMID: 26440996</ref>. | ||
| + | |||
| + | '''Quinohemoprotein alcohol dehydrogenase''' have pyrroloquinoline quinone (PQQ) as the prosthetic group <ref>PMID: 15234265</ref>. | ||
For chimeras of alcohol dehydrogenase see<br /> | For chimeras of alcohol dehydrogenase see<br /> | ||
| Line 20: | Line 34: | ||
*[[Q165E/S254K Double Mutant Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH]]<br /> | *[[Q165E/S254K Double Mutant Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH]]<br /> | ||
*[[Chimeras of alcohol dehydrogenases]]<br /> | *[[Chimeras of alcohol dehydrogenases]]<br /> | ||
| - | The <scene name='3fsr/Al1/2'>comparison</scene> of overall Cα backbone of all these chimeras (rmsd 0.45-0.65 Å) with those of the parent enzymes, did not reveal significant structural changes. So, the differences in the thermal stability of the chimeras and the parent enzymes could be caused by relatively small specific changes located at the important points of the NADP<sup>+</sup>-dependent alcohol dehydrogenases. For example see Cα superposition for the <font color='red'><b>X23<sub>(TET)</sub> chimera (red)</b></font> ([[3fpc]]) and its parent ADHs (<font color='blue'><b>TbADH, colored blue</b></font> ([[1ped]]), and < | + | The <scene name='3fsr/Al1/2'>comparison</scene> of overall Cα backbone of all these chimeras (rmsd 0.45-0.65 Å) with those of the parent enzymes, did not reveal significant structural changes. So, the differences in the thermal stability of the chimeras and the parent enzymes could be caused by relatively small specific changes located at the important points of the NADP<sup>+</sup>-dependent alcohol dehydrogenases. For example see Cα superposition for the <font color='red'><b>X23<sub>(TET)</sub> chimera (red)</b></font> ([[3fpc]]) and its parent ADHs (<font color='blue'><b>TbADH, colored blue</b></font> ([[1ped]]), and <span style="color:lime;background-color:black;font-weight:bold;">EhADH1, colored green</span> ([[1y9a]]). The [http://en.wikipedia.org/wiki/Root_mean_square_deviation RMSDs] of the TbADH−EhADH1, TbADH−Χ23<sub>(TET)</sub>, and EhADH1−Χ23<sub>(TET)</sub> were 0.68, 0.56, and 0.48 Å, respectively. |
<br/> | <br/> | ||
| - | {{TOC limit|limit=2}} | ||
{{Clear}} | {{Clear}} | ||
| - | |||
| - | ==Structure== | ||
| - | |||
| - | The initial scene (<scene name='Birrer_Sandbox_2/Whole_adh_molecule/3'>Domains of ADH</scene>) shows an overview of the molecule, allowing for a general look at the tertiary structure of alcohol dehydrogenase (it is complexed with Cl, Pyz, NAD, and Zn). A second scene (<scene name='Birrer_Sandbox_2/Close_look_at_ligand/2'>Closer Look at Subunit</scene>) shows a close view of the ligand within each subunit. Labels have been placed on NAD, CL, and Zn to clearly establish the structure. | ||
| - | |||
| - | |||
| - | Within alcohol dehydrogenase, <scene name='Birrer_Sandbox_2/The_active_site/1'>the active</scene> site of alcohol dehydrogenase has three important residues, Phe 93, Leu 57, and Leu 116. These three residues work together to bind to the alcohol substrate.<ref>''Protein: Alcohol Dehydrogenase''. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm></ref> | ||
| - | |||
| - | |||
| - | Zn plays an important role in the catalysis. It funtions by electrostatically stabilizing the oxygen in alcohol during the reaction, which causes the alcohol to be more acidic. At the <scene name='Birrer_Sandbox_2/Zinc_binding_site/1'>Zinc Binding Site</scene>, Zinc coordinates with Cys 146, Cys 174, and His 67.<ref>''Protein: Alcohol Dehydrogenase''. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm></ref> | ||
| - | |||
| - | |||
| - | NAD functions as a cosubstrate in the dehydration. NAD binds to numerous residues in a series of beta-alpha-beta folds. <scene name='Birrer_Sandbox_2/Nad_binding_site/1'>NAD Binding Region</scene> shows the domain where NAD binds, and many of the residues with which it interacts are selected. | ||
| - | <ref>''Protein: Alcohol Dehydrogenase''. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm></ref> | ||
| - | |||
| - | |||
| - | Alcohol dehydrogenase exists as a dimer with a zinc molecule complexed in each of the subunits. It has a SCOP catagory of an alpha and beta protein. At the N-terminal, there is a domain that is all beta; however, the C-Terminal domain is alpha and beta, so the catagory is alpha and beta. The C-Terminal core has 3 layers of alpha/beta/alpha and parallel beta sheets of 6 strands.<ref>''Protein: Alcohol dehydrogenase from Human (Homo sapiens), different isozymes''. SCOP. 2009. 1 March 2010 < http://scop.berkeley.edu/data/scop.b.d.c.b.b.c.html></ref> | ||
| - | |||
==Reaction and Mechanism== | ==Reaction and Mechanism== | ||
| Line 49: | Line 44: | ||
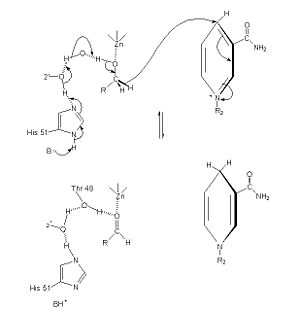
[[image:Mechanism of adh.jpg|left|300px]] | [[image:Mechanism of adh.jpg|left|300px]] | ||
{{Clear}} | {{Clear}} | ||
| - | The of alcohol dehydrogenase reaction is as follows: CH3CH2OH + NAD+ -> CH3COH (acetaldehyde) + NADH + H+ (Note: The reaction is actually reversible although the arrow does not show it) <ref>Voet, et. al. ''Fundamentals of Biochemistry: 3rd Edition''. Hoboken: Wiley & Sons, Inc, 2008.</ref> The step-wise reduction mechanism for ADH is shown on the left. In the mechanism, His 51 is deprotonated and activated by a base catalyst. This allows histidine to accept a proton from NAD, which also draws a proton Thr 48. As a result of the proton transfer, the Thr is prepared to accept a proton from the alcohol substrate. While Thr accepts the proton, there is also a hydride transfer to NAD. The whole process can be summarized as the oxidation of an alcohol to an aldehyde in concert with the transfer of a hydride to NAD.<ref>''Protein: Alcohol Dehydrogenase''. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm></ref><ref>''Protein: Alcohol Dehydrogenase''. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm></ref> | + | The of alcohol dehydrogenase reaction is as follows: CH3CH2OH + NAD+ -> CH3COH (acetaldehyde) + NADH + H+ (Note: The reaction is actually reversible although the arrow does not show it) <ref>Voet, et. al. ''Fundamentals of Biochemistry: 3rd Edition''. Hoboken: Wiley & Sons, Inc, 2008.</ref> The step-wise reduction mechanism for ADH is shown on the left. In the mechanism, His 51 is deprotonated and activated by a base catalyst. This allows histidine to accept a proton from NAD, which also draws a proton Thr 48. As a result of the proton transfer, the Thr is prepared to accept a proton from the alcohol substrate. While Thr accepts the proton, there is also a hydride transfer to NAD. The whole process can be summarized as the oxidation of an alcohol to an aldehyde in concert with the transfer of a hydride to NAD.<ref>''Protein: Alcohol Dehydrogenase''. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://web.archive.org/web/20080307193453/http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm></ref><ref>''Protein: Alcohol Dehydrogenase''. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://web.archive.org/web/20080307193453/http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm></ref> |
The Mechanism for alcohol dehydrogenase follows an random bisubstrate mechanism.<ref>Voet, et. al. ''Fundamentals of Biochemistry: 3rd Edition''. Hoboken: Wiley & Sons, Inc, 2008.</ref> In the mechanism, the NAD+ and alcohol bind to the enzyme, so that the enzyme is now attached to the two subtrates. While attached, the hydrogen is formally transferred from the alcohol to NAD, resulting in the products NADH and a ketone or aldehyde. The two products are then released, and the enzyme has catalyzed the reaction. | The Mechanism for alcohol dehydrogenase follows an random bisubstrate mechanism.<ref>Voet, et. al. ''Fundamentals of Biochemistry: 3rd Edition''. Hoboken: Wiley & Sons, Inc, 2008.</ref> In the mechanism, the NAD+ and alcohol bind to the enzyme, so that the enzyme is now attached to the two subtrates. While attached, the hydrogen is formally transferred from the alcohol to NAD, resulting in the products NADH and a ketone or aldehyde. The two products are then released, and the enzyme has catalyzed the reaction. | ||
| Line 67: | Line 62: | ||
{{Clear}} | {{Clear}} | ||
| - | The double [http://en.wikipedia.org/wiki/Mutation mutant] of the chimera Χ21<sub>(TCT)</sub> (cofactor-binding domain of thermophilic TbADH replaced by that of mesophilic CbADH) Q165E/S254K-X21<sub>(TCT)</sub> ([[3ftn]]) was constructed by [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis]. In both TbADH and CbADH, Lys257 and Asp237 form an intrasubunit ion pair, in TbADH, Asp237 is also involved in an ion pair bridge with Arg304 of the adjacent monomer. In addition, Arg304 forms intersubunit salt bridge with Glu165 of the first monomer. Therefore, a <scene name='3fsr/Al/2'>four-member ion pair network</scene> involving Lys257, Asp237, and Glu165 of one monomer and Arg304 of the adjacent one is present in TbADH (the names of monomers are in brackets). However in mesophilic CbADH (and, therefore, in the chimera Χ21<sub>(TCT)</sub>, [[3fsr]]) the Gln is situated in position 165 (instead Glu of TbADH) and Met in position 304 (instead Arg of TbADH), so, such an ion pair network does not exist. In the double mutant Q165E/S254K-X21<sub>(TCT)</sub> reverse mutation Q165E reconstructs this network (as in parent thermophilic TbADH) that led to significant enhancement of the thermal stability of CbADH (ΔT<sub>1/2</sub><sup>60 min</sup> = 5.4 °C). <font color='magenta'><b>Chimera X21<sup>(TCT)</sup> ([[3fsr]]) is colored magenta</b></font> and < | + | The double [http://en.wikipedia.org/wiki/Mutation mutant] of the chimera Χ21<sub>(TCT)</sub> (cofactor-binding domain of thermophilic TbADH replaced by that of mesophilic CbADH) Q165E/S254K-X21<sub>(TCT)</sub> ([[3ftn]]) was constructed by [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis]. In both TbADH and CbADH, Lys257 and Asp237 form an intrasubunit ion pair, in TbADH, Asp237 is also involved in an ion pair bridge with Arg304 of the adjacent monomer. In addition, Arg304 forms intersubunit salt bridge with Glu165 of the first monomer. Therefore, a <scene name='3fsr/Al/2'>four-member ion pair network</scene> involving Lys257, Asp237, and Glu165 of one monomer and Arg304 of the adjacent one is present in TbADH (the names of monomers are in brackets). However in mesophilic CbADH (and, therefore, in the chimera Χ21<sub>(TCT)</sub>, [[3fsr]]) the Gln is situated in position 165 (instead Glu of TbADH) and Met in position 304 (instead Arg of TbADH), so, such an ion pair network does not exist. In the double mutant Q165E/S254K-X21<sub>(TCT)</sub> reverse mutation Q165E reconstructs this network (as in parent thermophilic TbADH) that led to significant enhancement of the thermal stability of CbADH (ΔT<sub>1/2</sub><sup>60 min</sup> = 5.4 °C). <font color='magenta'><b>Chimera X21<sup>(TCT)</sup> ([[3fsr]]) is colored magenta</b></font> and <span style="color:cyan;background-color:black;font-weight:bold;">the double mutant Q165E/S254K-X21<sup>(TCT)</sup> cyan</span> ([[3ftn]]). In chimera X21<sub>(TCT)</sub>, position 254 is occupied by Ser (due to sequence of exchanged domain). The replacement of Ser254 of CbADH with Lys significantly enhances the stability of the enzyme, due to the formation of <scene name='3fsr/Al/3'>intrasubunit Lys254 and Glu280 ion pair</scene>. However, this replacing of Ser254 by Lys had a negligible effect on the thermal stability, in contrast to mutation Q165E mentioned above. |
{{Clear}} | {{Clear}} | ||
| - | The <scene name='3fsr/Al1/2'>comparison</scene> of overall Cα backbone of all these chimeras (rmsd 0.45-0.65 Å) with those of the parent enzymes, did not reveal significant structural changes. So, the differences in the thermal stability of the chimeras and the parent enzymes could be caused by relatively small specific changes located at the important points of the NADP<sup>+</sup>-dependent alcohol dehydrogenases. For example see Cα superposition for the <font color='red'><b>X23<sub>(TET)</sub> chimera (red)</b></font> ([[3fpc]]) and its parent ADHs (<font color='blue'><b>TbADH, colored blue</b></font> ([[1ped]]), and < | + | The <scene name='3fsr/Al1/2'>comparison</scene> of overall Cα backbone of all these chimeras (rmsd 0.45-0.65 Å) with those of the parent enzymes, did not reveal significant structural changes. So, the differences in the thermal stability of the chimeras and the parent enzymes could be caused by relatively small specific changes located at the important points of the NADP<sup>+</sup>-dependent alcohol dehydrogenases. For example see Cα superposition for the <font color='red'><b>X23<sub>(TET)</sub> chimera (red)</b></font> ([[3fpc]]) and its parent ADHs (<font color='blue'><b>TbADH, colored blue</b></font> ([[1ped]]), and <span style="color:lime;background-color:black;font-weight:bold;">EhADH1, colored green</span> ([[1y9a]]). The [http://en.wikipedia.org/wiki/Root_mean_square_deviation RMSDs] of the TbADH−EhADH1, TbADH−Χ23<sub>(TET)</sub>, and EhADH1−Χ23<sub>(TET)</sub> were 0.68, 0.56, and 0.48 Å, respectively. |
{{Clear}} | {{Clear}} | ||
| - | The 3D structure of CbADH with the substitution Q100P (<scene name='2b83/Tet/3'>tetramer</scene>) was solved at 2.25 Å resolution ([[2b83]]). The <scene name='2b83/Mut/1'>substitution</scene> of Gln100 with Pro did not cause significant structural changes in the protein structure. The residues of the < | + | The 3D structure of CbADH with the substitution Q100P (<scene name='2b83/Tet/3'>tetramer</scene>) was solved at 2.25 Å resolution ([[2b83]]). The <scene name='2b83/Mut/1'>substitution</scene> of Gln100 with Pro did not cause significant structural changes in the protein structure. The residues of the <span style="color:lime;background-color:black;font-weight:bold;">wildtype protein are colored green</span> and the residues of the <span style="color:cyan;background-color:black;font-weight:bold;">mutant one in cyan</span>. Only [http://en.wikipedia.org/wiki/Hydrogen_bond 2 H-bonds] were lost, one between Oε1 of Gln100 and the main chain N of Gly297, and the second between Nε2 of Gln100 and the main chain carbonyl O of Gly297. The mutation caused that an additional CH<sub>2</sub> group (Cδ of Pro100) is surrounded by nonpolar residues: Pro88 (3.8 Å), Trp90 (3.5 Å), and Val95 (4 Å). These residues (P100, P88, W90, and V95) are situated on a protruding lobe of the protein. An additional 11 [http://en.wikipedia.org/wiki/Aliphatic_compound aliphatic] and [http://en.wikipedia.org/wiki/Aromatic aromatic] carbon atoms are situated within the distance of 6 Å from Cδ of Pro100 (two [http://en.wikipedia.org/wiki/Methyl_group methyl groups] of Val95; three carbon atoms of the Trp90 [http://en.wikipedia.org/wiki/Indole indole] group; Cβ and Cγ [http://en.wikipedia.org/wiki/Methylene methylene] groups of Pro100; Cβ and Cγ of Gln101, and two carbons of the Phe99 [http://en.wikipedia.org/wiki/Phenyl_group phenyl] ring). |
{{Clear}} | {{Clear}} | ||
| - | Ribbon diagram of the EhADH1 <scene name='2oui/Tet/1'>tetramer</scene> ([[2oui]]). Proline residues (ball representation) are colored < | + | Ribbon diagram of the EhADH1 <scene name='2oui/Tet/1'>tetramer</scene> ([[2oui]]). Proline residues (ball representation) are colored <span style="color:orange;background-color:black;font-weight:bold;">orange (Pro275)</span> (which is important for thermal stabilization) and <span style="color:cyan;background-color:black;font-weight:bold;">cyan (Pro100)</span>. <scene name='2oui/Tet/5'>Superposition</scene> of the structures of the <span style="color:lime;background-color:black;font-weight:bold;">wild-type apo-EhADH1 (colored green</span>, [[1y9a]]) and the <span style="color:orange;background-color:black;font-weight:bold;">apo D275P-EhADH1 mutant (colored orange)</span> ([[2oui]]). <font color='red'><b>Pro275 and Asp275 are labeled red.</b></font> Residues within a distance of 4 Å from the mutation are shown (names of monomers are in brackets). Replacing <scene name='2oui/Tet/8'>Asp275</scene> with <scene name='2oui/Tet/7'>Pro</scene> significantly enhanced the thermal stability of EhADH1: ΔT<sub>1/2</sub><sup>60min</sup> = +9.3°C, ΔT<sub>1/2</sub><sup>CD</sup> = +10°C. The reverse mutation in the thermophilic <scene name='Tetrameric_alcohol_dehydrogenases/Mut/3'>TbADH</scene> ([[1ykf]]; <font color='magenta'><b>colored magenta</b></font>) - substitution of wt TbADH Pro275 with <scene name='Tetrameric_alcohol_dehydrogenases/Mut/2'>Asp</scene> ([[2nvb]]; <span style="color:cyan;background-color:black;font-weight:bold;">colored cyan</span>) reduced the thermal stability of the enzyme: ΔT<sub>1/2</sub><sup>60min</sup> = -13.8°C, ΔT<sub>1/2</sub><sup>CD</sup> = -18.8°C. Nitrogen and oxygen atoms are colored in [http://en.wikipedia.org/wiki/CPK_coloring CPK colors]. <font color='red'><b>Pro275 and Asp275 are labeled red</b></font> (names of monomers are in brackets). These findings indicate that a single proline mutation is responsible for the significant differences in the thermal stability of ADHs, and show the importance of prolines in the protein stability. It was also shown that substitution by proline at the important positions could significantly stabilize the protein.<ref>PMID 17063493</ref><ref>PMID 18260103</ref><ref>PMID 20102159</ref> |
{{Clear}} | {{Clear}} | ||
| - | </StructureSection> | ||
| - | __NOTOC__ | ||
| - | ==Additional Resources== | ||
| - | For additional information, see: [[Carbohydrate Metabolism]]<br /> | ||
| - | |||
| - | == 3D Structures of Alcohol dehydrogenase== | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | |||
| - | ===ADH I=== | ||
| - | |||
| - | [[3jv7]] – RrADH I – ''Rhodococcus rubber''<br /> | ||
| - | [[2vna]] - hADH I catalytic domain - human<br /> | ||
| - | [[2hcy]] – yADH I – yeast<br /> | ||
| - | [[4eex]] – LlADH I – ''Lactococcus lactis''<br /> | ||
| - | [[4eez]] – LlADH I (mutant) | ||
| - | |||
| - | ''ADH I binary complex'' | ||
| - | |||
| - | [[1u3t]] – hADH I α chain + inhibitor<br /> | ||
| - | [[1hsz]], [[1hdz]], [[3hud]] - hADH I β chain + NAD<br /> | ||
| - | [[1u3w]] - hADH I γ chain + inhibitor<br /> | ||
| - | [[1ht0]] - hADH I γ chain (mutant) + NAD | ||
| - | |||
| - | ''ADH I ternary complex'' | ||
| - | |||
| - | [[2xaa]] – RrADH I + NAD + alcohol<br /> | ||
| - | [[3fx4]] – pADH I + NADP + inhibitor – pig<br /> | ||
| - | [[2w98]], [[2w4q]] – hADH I catalytic domain + NADP + inhibitor<br /> | ||
| - | [[1hso]] - hADH I α chain + NAD + pyrazole derivative<br /> | ||
| - | [[1hdx]] - hADH I β chain + NAD + alcohol<br /> | ||
| - | [[1u3u]], [[1u3v]] - hADH I β chain + inhibitor<br /> | ||
| - | [[1deh]], [[1hdy]] - hADH I β chain + NAD + pyrazole derivative<br /> | ||
| - | [[1htb]] - hADH I β3 chain + NAD + pyrazole derivative | ||
| - | |||
| - | ===ADH II=== | ||
| - | |||
| - | [[3owo]] – ZmADH II iron-dependent – ''Zymomonas mobilis'' | ||
| - | |||
| - | ''ADH II binary complex'' | ||
| - | |||
| - | [[3ox4]] - ZmADH II iron-dependent + NAD<br /> | ||
| - | [[3cos]] - hADH II + NAD + Zn<br /> | ||
| - | [[1e3e]] – mADH II + NADH – mouse<br /> | ||
| - | [[1e3l]] - mADH II (mutant) + NADH<br /> | ||
| - | [[1e3i]] - mADH II + NADH + inhibitor | ||
| - | |||
| - | '''ADH III''' | ||
| - | |||
| - | [[1m6h]], [[1m6w]], [[1teh]] - hADH III χ chain<br /> | ||
| - | [[2fze]] - hADH III χ chain + ADP-ribose<br /> | ||
| - | [[2fzw]], [[1mp0]] - hADH III χ chain + NAD<br /> | ||
| - | [[1mc5]] – hADH III χ chain + glutathione + NADH<br /> | ||
| - | [[1ma0]] - hADH III χ chain + dodecanoic acid + NAD<br /> | ||
| - | [[3qj5]] - hADH III χ chain + inhibitor + NAD<br /> | ||
| - | [[4dl9]], [[4dlb]] – tADH III + NAD – tomato<br /> | ||
| - | [[4dla]] – tADH III | ||
| - | |||
| - | ===ADH IV=== | ||
| - | |||
| - | [[1ye3]], [[8adh]], [[5adh]] - hoADH IV e chain – horse<br /> | ||
| - | [[1qlj]] - hoADH IV e chain (mutant) <br /> | ||
| - | [[3iv7]] – ADH IV – ''Corynebacterium glutamicum'' | ||
| - | |||
| - | ''ADH IV binary complex'' | ||
| - | |||
| - | [[2jhf]], [[2jhg]], [[1het]], [[1heu]], [[1hf3]], [[1ee2]], [[2oxi]], [[2ohx]], [[6adh]] - hoADH IV e chain + NAD<br /> | ||
| - | [[1adb]], [[1adc]], [[1adf]], [[1adg]], [[7adh]] - hoADH IV e chain + NAD derivative<br /> | ||
| - | [[1mgo]], [[1ju9]], [[1qlh]], [[1a72]] - hoADH IV e chain (mutant) + NAD<br /> | ||
| - | [[1d1s]], [[1agn]] – hADH IV σ chain + NAD<br /> | ||
| - | [[1d1t]] - hADH IV σ chain (mutant) + NAD | ||
| - | |||
| - | |||
| - | ''ADH IV ternary complex'' | ||
| - | |||
| - | [[3oq6]], [[1qv6]], [[1qv7]], [[1a71]], [[1axe]], [[1axg]], [[4nfh]], [[4nfs]], [[4ng5]] – hoADH IV e chain (mutant) + NAD + alcohol<br /> | ||
| - | [[4dwv]], [[4dxh]] - hoADH IV e chain + NAD + alcohol<br /> | ||
| - | [[1p1r]], [[1ldy]], [[1lde]] - hoADH IV e chain + NADH + formamide derivative<br /> | ||
| - | [[1n92]] - hoADH IV e chain + NAD + pyrazole derivative<br /> | ||
| - | [[1bto]], [[3bto]] - hoADH IV e chain + NADH + butylthiolane derivative<br /> | ||
| - | [[1n8k]] - hoADH IV e chain (mutant) + NAD + pyrazole<br /> | ||
| - | [[1mg0]], [[1hld]] - hoADH IV e chain + NAD + alcohol<br /> | ||
| - | |||
| - | ===ADH=== | ||
| - | |||
| - | [[1a4u]] – SlADH – ''Scaptodrosophila lebanonensis''<br /> | ||
| - | [[3my7]] – ADH ACDH domain – ''Vibrio parahaemolyticus''<br /> | ||
| - | [[3meq]] – ADH – ''Brucella suis''<br /> | ||
| - | [[3l4p]] – ADH – ''Desulfovibrio gigas''<br /> | ||
| - | [[1jvb]] - SsADH – ''Sulfolobus solfataricus''<br /> | ||
| - | [[3i4c]], [[1nto]], [[1nvg]] – SsADH (mutant) <br /> | ||
| - | [[3goh]] – ADH – ''Shewanella oneidensis''<br /> | ||
| - | [[3gaz]] – ADH residues 2-334 – ''Novosphingobium aromaticivorans''<br /> | ||
| - | [[2eih]] – ADH – ''Thermus thermophilus''<br /> | ||
| - | [[1rjw]] – GsADH – ''Geobacillus stearothermophilus''<br /> | ||
| - | [[1vj0]], [[1vhd]] – TmADH -''Thermotoga maritima''<br /> | ||
| - | [[2eer]] – ADH – ''Sulfolobus tokodaii''<br /> | ||
| - | [[3uog]] – ADH – ''Sinorhizobium meliloti'' | ||
| - | |||
| - | ''ADH binary complex'' | ||
| - | |||
| - | [[3l77]], [[3tn7]] – ADH short-chain + NADP – ''Thermococcus sibiricus''<br /> | ||
| - | [[1h2b]] – ADH + NAD – ''Aeropyrum pernix''<br /> | ||
| - | [[1f8f]] – Benzyl-ADH + NAD – ''Acinetobacter calcoaceticus''<br /> | ||
| - | [[1o2d]] - TmADH + NADP <br /> | ||
| - | [[3ip1]] – TmADH + Cd<br /> | ||
| - | [[1b16]], [[1b14]], [[1b15]] - SlADH + NAD derivative<br /> | ||
| - | [[1cdo]] – ADH + NAD - cod<br /> | ||
| - | [[1rhc]] – ADH F420-dependent +F420-acetone – ''Methanoculleus thermophilus''<br /> | ||
| - | [[3s2e]] – ReADH + NAD + Zn<br /> | ||
| - | [[1agn]] – hADH (sigma) +NAD<br /> | ||
| - | [[3pii]] – GsADH + butyramide<br /> | ||
| - | [[3rj5]], [[3rj9]] – SlADH (mutant) + NAD<br /> | ||
| - | [[3s1l]] – ReADH + Zn – ''Ralstonia eutropha''<br /> | ||
| - | [[3jzd]] – ReADH + NAD<br /> | ||
| - | |||
| - | ''ADH ternary complex'' | ||
| - | |||
| - | [[1mg5]] – ADH + NADH + acetate – ''Drosophila melanogaster''<br /> | ||
| - | [[1r37]] – SsADH + NAD + alcohol<br /> | ||
| - | [[1sby]] – SlADH + NAD + alcohol<br /> | ||
| - | [[1b2l]] - SlADH + NAD + cyclohexanone<br /> | ||
| - | [[1llu]] - ADH + NAD + alcohol – ''Pseudomonas aeruginosa''<br /> | ||
| - | [[3cv7]] – pADH + NAD + NAP<br /> | ||
| - | [[3rf7]] – SoADH + NAD + Fe + Ni<br /> | ||
| - | [[3s2e]] – ReADH + NAD + Zn<br /> | ||
| - | [[3s2f]], [[3s2g]] – ReADH + NAD + Zn + furfural<br /> | ||
| - | [[4gkv]] – ADH + NAD + Zn + peptide – ''Escherichia coli''<br /> | ||
| - | [[4jji]], [[4gl4]], [[3uko]] - AtADH III + NAD + Zn – ''Arabidopsis thaliana'' <br /> | ||
| - | [[4l0q]] - AtADH III (mutant) + NAD + Zn <br /> | ||
| - | |||
| - | ===NADP-dependent ADH=== | ||
| - | |||
| - | [[1ped]] - CbADH – ''Clostridium beijerinckii''<br /> | ||
| - | [[2b83]], [[1jqb]] – CbADH (mutant) <br /> | ||
| - | [[2nvb]] - TbADH (mutant) – ''Thermoanaerobacter brockii''<br /> | ||
| - | [[3ftn]], [[3fpc]], [[3fpl]], [[3fsr]] – ADH chimera<br /> | ||
| - | [[1y9a]] - EhADH – ''Entamoeba histolytica''<br /> | ||
| - | [[2oui]] – EhADH (mutant) <br /> | ||
| - | [[1p0c]] – RpADH8 – ''Rana perezi'' <br /> | ||
| - | [[4hfj]] – toADH – tobacco<br /> | ||
| - | [[4gac]] - mADH | ||
| - | |||
| - | ''NADP-dependent ADH binary complex'' | ||
| - | |||
| - | [[1kev]] – CbADH + NADPH<br /> | ||
| - | [[1bxz]] – CbADH catalytic domain + alcohol<br /> | ||
| - | [[1ykf]] – TbADH + NADP<br /> | ||
| - | [[3h4g]] – pADH + NADP<br /> | ||
| - | [[1p0f]] – RpADH + NADP<br /> | ||
| - | [[4hfm]] - toADH + NADP<br /> | ||
| - | [[4hfn]] - toADH + NADP + coniferaldehyde<br /> | ||
| - | [[4jbg]] - PaADH + Zn – ''Pyrobaculum aerophilum''<br /> | ||
| - | [[4jbh]] - PaADH + Zn + Co<br /> | ||
| - | [[4jbi]] - PaADH + NADP + Zn<br /> | ||
| - | |||
| - | ===R-specific ADH=== | ||
| - | |||
| - | [[1nxq]] - LbRADH – ''Lactobacillus brevis''<br /> | ||
| - | [[1zk2]], [[1zk3]] - LbRADH (mutant)<br /> | ||
| - | [[1zjy]], [[1zjz]], [[1zk0]], [[1zk1]] – LbRADH (mutant) + NADH + alcohol<br /> | ||
| - | [[1zk4]] - LbRADH (mutant) + NADH + acetophenone | ||
| - | |||
| - | ===Specific alcohol ADH=== | ||
| - | |||
| - | [[2cf5]], [[2cf6]] – Cinnamyl-AtADH <br /> | ||
| - | [[1piw]], [[1q1n]], [[1ps0]] – Cinnamyl-yADH<br /> | ||
| - | [[3two]] - Cinnamyl-ADH + NADP – ''Helicobacter pylori''<br /> | ||
| - | [[1m2w]] – Mannitol-ADH – ''Pseudomonas fluorescens'' <br /> | ||
| - | [[1w6s]] – Methanol-ADH – ''Methylobacterium extorquens''<br /> | ||
| - | [[1yqx]] – Sinapyl-aADH II – aspen<br /> | ||
| - | [[1yqd]] – Sinapyl-aADH II + NADP<br /> | ||
| - | [[1bdb]] – Biphenyl dihydrodiol-ADH + NAD - ''Pseudomonas'' | ||
| - | |||
| - | ===Quinohemoprotein ADH=== | ||
| - | |||
| - | [[1kv9]], [[1yiq]] – PpQADH II + PQQ + heme – ''Pseudomonas putida''<br /> | ||
| - | [[1kb0]] - QADH I + PQQ + heme – ''Comamonas testosteroni'' | ||
| - | |||
| - | ===Hydroxyacyl-CoA dehydrogenase=== | ||
| - | |||
| - | ''Short chain HADH'' | ||
| - | |||
| - | [[1so8]] – hSHCDH II – human<BR /> | ||
| - | [[3rqs]] - hSHCDH <BR /> | ||
| - | [[1f14]] - hSHCDH (mutant) | ||
| - | |||
| - | ''Short chain HADH binary complex'' | ||
| - | |||
| - | [[1f12]] - hSHCDH (mutant) + hydroxybutyryl-CoA<BR /> | ||
| - | [[1f17]], [[1lsj]], [[1lso]] - hSHCDH (mutant) + NAD<BR /> | ||
| - | [[1zbq]] - hSHCDH IV + NAD<BR /> | ||
| - | [[1e3s]] - rSHCDH + NAD – rat | ||
| - | |||
| - | ''Short chain HADH ternary complex'' | ||
| - | |||
| - | [[1u7t]] - hSHCDH II + inhibitor + NAD<BR /> | ||
| - | [[1f0y]] - hSHCDH + acetoacetyl-CoA + NAD<BR /> | ||
| - | [[1il0]], [[1m75]], [[1m76]] - hSHCDH (mutant) + acetoacetyl-CoA + NAD<BR /> | ||
| - | [[1e3w]] - rSHCDH + 3-keto-butyrate + NAD<BR /> | ||
| - | [[1e6w]] - rSHCDH + estradiol + NAD<BR /> | ||
| - | |||
| - | ''Unspecified HADH'' | ||
| - | |||
| - | [[1uay]] - HADH II – ''Thermus thermophilus''<BR /> | ||
| - | [[1zej]], [[3ctv]] - HADH – ''Archaeoglobus fulgidus''<BR /> | ||
| - | [[1zcj]] - rHADH <BR /> | ||
| - | [[2x58]] - rHADH + CoA <BR /> | ||
| - | [[2et6]] – HADH (mutant) – ''Candida tropicalis'' | ||
| + | == 3D Structures of alcohol dehydrogenase== | ||
| + | [[Alcohol dehydrogenase 3D structures]] | ||
| + | </StructureSection> | ||
| + | ==Additional Resources== | ||
| + | For additional information, see: | ||
| + | *[[Carbohydrate Metabolism]] | ||
| + | *[[Horse Liver Alcohol Dehydrogenase]] | ||
==References== | ==References== | ||
Current revision
| |||||||||||
Additional Resources
For additional information, see:
References
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Sutak R, Hrdy I, Dolezal P, Cabala R, Sedinova M, Lewin J, Harant K, Muller M, Tachezy J. Secondary alcohol dehydrogenase catalyzes the reduction of exogenous acetone to 2-propanol in Trichomonas vaginalis. FEBS J. 2012 Aug;279(15):2768-80. doi: 10.1111/j.1742-4658.2012.08661.x. Epub, 2012 Jul 9. PMID:22686835 doi:http://dx.doi.org/10.1111/j.1742-4658.2012.08661.x
- ↑ Ismaiel AA, Zhu CX, Colby GD, Chen JS. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J Bacteriol. 1993 Aug;175(16):5097-105. PMID:8349550
- ↑ Rozeboom HJ, Yu S, Mikkelsen R, Nikolaev I, Mulder HJ, Dijkstra BW. Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenes. A versatile dehydrogenase oxidizing alcohols and carbohydrates. Protein Sci. 2015 Oct 6. doi: 10.1002/pro.2818. PMID:26440996 doi:http://dx.doi.org/10.1002/pro.2818
- ↑ Toyama H, Mathews FS, Adachi O, Matsushita K. Quinohemoprotein alcohol dehydrogenases: structure, function, and physiology. Arch Biochem Biophys. 2004 Aug 1;428(1):10-21. PMID:15234265 doi:10.1016/j.abb.2004.03.037
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://web.archive.org/web/20080307193453/http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://web.archive.org/web/20080307193453/http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Bille V, Remacle J. Simple-kinetic descriptions of alcohol dehydrogenase after immobilization on tresyl-chloride-activated agarose. Eur J Biochem. 1986 Oct 15;160(2):343-8. PMID:3769934
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Blomstrand R, Ostling-Wintzell H, Lof A, McMartin K, Tolf BR, Hedstrom KG. Pyrazoles as inhibitors of alcohol oxidation and as important tools in alcohol research: an approach to therapy against methanol poisoning. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3499-503. PMID:115004
- ↑ Alcohol Dehydrogenase. Worthington Biochemical Corporation . 31 March 2010 < http://http://www.worthington-biochem.com/ADH/default.html>
- ↑ Alcohol Dehydrogenase.Worthington Biochemical Corporation . 31 March 2010 < http://http://www.worthington-biochem.com/ADH/default.html>
- ↑ Goihberg E, Dym O, Tel-Or S, Levin I, Peretz M, Burstein Y. A single proline substitution is critical for the thermostabilization of Clostridium beijerinckii alcohol dehydrogenase. Proteins. 2007 Jan 1;66(1):196-204. PMID:17063493 doi:10.1002/prot.21170
- ↑ Goihberg E, Dym O, Tel-Or S, Shimon L, Frolow F, Peretz M, Burstein Y. Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution. Proteins. 2008 Feb 7;. PMID:18260103 doi:10.1002/prot.21946
- ↑ Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon L, Frolow F, Burstein Y. Biochemical and Structural Properties of Chimeras Constructed by Exchange of Cofactor-Binding Domains in Alcohol Dehydrogenases from Thermophilic and Mesophilic Microorganisms. Biochemistry. 2010 Feb 9. PMID:20102159 doi:10.1021/bi901730x
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, David Birrer