Insulin
From Proteopedia
| (35 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:Insulin.gif|right|Insulin chain A and B. For interactive figures, scroll down and click on green links]] | + | [[Image:Insulin.gif|right|frame|Insulin chain A and B. For interactive figures, scroll down and click on green links]] |
| - | '''Insulin''' is a peptide hormone that helps to maintain blood sugar within a healthy range by regulating [[Carbohydrate Metabolism|carbohydrate]] and lipid metabolism throughout the body | + | '''Insulin''' is a peptide [[hormone]] that helps to maintain blood sugar within a healthy range by regulating [[Carbohydrate Metabolism|carbohydrate]] and lipid metabolism throughout the body. It is secreted by specialized cells in the pancreas and acts by binding to [[insulin receptor|insulin receptors]] on other cells. Insulin in its mature form contains two peptide chains connected by [[Cystine|disulfide crosslinks]], and occurs either as monomer or as hexamer. Administering insulin in carefully determined doses at the appropriate times is used in managing [[diabetes]], a chronic condition where the body fails to maintain blood sugar levels by itself. |
| + | *'''Proinsulin''' is the insulin precursor which is converted in the pancreas to insulin and C-peptide<ref>PMID:403392</ref>. | ||
| + | *'''Leginsulin''' is an insulin-like peptide found in legumes <ref>PMID:30442953</ref>. | ||
| + | *'''Insulin lispro''' is recombinant insulin which has more rapid absorption than regular insulin<ref>PMID:9339963</ref>. See also [[Insulin lispro]]. | ||
| + | *'''Insulin glargine''' is a long-acting insulin <ref>PMID:10730548</ref>. See also [[Insulin glargine]]. | ||
==Other Proteopedia pages about or relating to insulin== | ==Other Proteopedia pages about or relating to insulin== | ||
| Line 8: | Line 12: | ||
[[Diabetes & Hypoglycemia]]<br> | [[Diabetes & Hypoglycemia]]<br> | ||
[[Insulin (Hebrew)]]<br> | [[Insulin (Hebrew)]]<br> | ||
| - | [[Insulin | + | [[Insulin aspart]]<br> |
[[Insulin_mo-or-sl|Insulin and pro-insulin (Hebrew)]]<br> | [[Insulin_mo-or-sl|Insulin and pro-insulin (Hebrew)]]<br> | ||
[[Insulin-Degrading Enzyme]]<br> | [[Insulin-Degrading Enzyme]]<br> | ||
[[Molecular_Playground/Insulin]]<br> | [[Molecular_Playground/Insulin]]<br> | ||
[[Insulin Structure & Function]]<br> | [[Insulin Structure & Function]]<br> | ||
| - | [[Insulin | + | [[Insulin signal transduction pathway]]<br> |
'''Proteins that interact with insulin''' | '''Proteins that interact with insulin''' | ||
| Line 21: | Line 25: | ||
[[Insulin receptor]]<br> | [[Insulin receptor]]<br> | ||
[[Insulin-like growth factor receptor]]<br> | [[Insulin-like growth factor receptor]]<br> | ||
| + | |||
'''Tutorials we would love to see''' | '''Tutorials we would love to see''' | ||
| Line 30: | Line 35: | ||
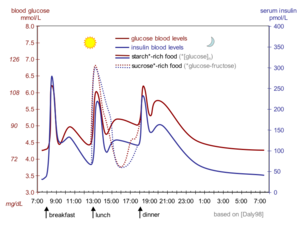
[[Image:Glucose insulin day.png|300px|left|thumb|Schematic of daily changes of blood sugar (red) and insulin (blue)]] | [[Image:Glucose insulin day.png|300px|left|thumb|Schematic of daily changes of blood sugar (red) and insulin (blue)]] | ||
| - | Our blood sugar level (i.e. glucose concentration) varies over time with food intake and exercise, but has to remain in a narrow range so we stay healthy (not become hyperglycemic or hypoglycemic). Insulin, together with [[glucagon]], regulates blood sugar levels by changing fuel metabolism in all cells <ref>https://www.yourhormones.info/hormones/insulin/</ref> | + | Our blood sugar level (i.e. glucose concentration) varies over time with food intake and exercise, but has to remain in a narrow range so we stay healthy (not become hyperglycemic or hypoglycemic). Insulin, together with [[glucagon]], regulates blood sugar levels by changing fuel metabolism in all metabolic cells (e.g. liver, fat, kidney, muscle and nerve cells) on a timescale of minutes and hours<ref>https://www.yourhormones.info/hormones/insulin/</ref><ref>PMID:10927996</ref><ref>DOI:10.1111/dom.13402</ref>. In simplest terms, the presence of insulin in the blood signals the well-fed stage, while the presence of glucagon signals the fasting stage. |
Biosynthesis and processing of insulin occurs in the [https://en.wikipedia.org/wiki/Beta_cell beta cells] of the pancreas. The beta cells are found in the islets of Langerhans, which also contain the alpha cells that synthesize glucagon. Insulin is made as a 110 amino acid pre-proinsulin, which is processed to the mature 51 amino acid insulin and targeted to secretory vesicles. In a healthy adult, about 200 units of insulin are available in the pancreas, of which 30-50 units are secreted daily<ref>https://www.britannica.com/science/insulin</ref>. With a unit of insulin corresponding to 0.0347 mg mature insulin <ref>https://www.who.int/biologicals/expert_committee/BS_2143_Human_Recombinant_Insulin_final.pdf</ref>, the body contains only about 7 mg of insulin. In its stored form, insulin is a hexamer complexed to zinc ions. | Biosynthesis and processing of insulin occurs in the [https://en.wikipedia.org/wiki/Beta_cell beta cells] of the pancreas. The beta cells are found in the islets of Langerhans, which also contain the alpha cells that synthesize glucagon. Insulin is made as a 110 amino acid pre-proinsulin, which is processed to the mature 51 amino acid insulin and targeted to secretory vesicles. In a healthy adult, about 200 units of insulin are available in the pancreas, of which 30-50 units are secreted daily<ref>https://www.britannica.com/science/insulin</ref>. With a unit of insulin corresponding to 0.0347 mg mature insulin <ref>https://www.who.int/biologicals/expert_committee/BS_2143_Human_Recombinant_Insulin_final.pdf</ref>, the body contains only about 7 mg of insulin. In its stored form, insulin is a hexamer complexed to zinc ions. | ||
| Line 36: | Line 41: | ||
Insulin is constantly released into the blood stream at a basal level. When blood glucose levels rise after a meal containing carbohydrates (sugars and starches), increased intracellular glucose in the beta cells triggers a series of processes that change the intracellular calcium concentration. First, the calcium concentration rises and stays elevated for 5-10 minutes, and then it spikes in intervals of roughly 3 minutes. Whenever calcium concentrations are high, insulin-containing vesicles merge with the plasma membrane, releasing insulin into the blood stream in the same temporal pattern as the calcium spikes<ref>DOI: 10.1007/s00125-009-1314-y</ref>. The insulin is released into the bloodstream, where it first reaches the liver via the portal vein<ref>"https://www.diabetesselfmanagement.com/diabetes-resources/definitions/portal-vein/"</ref>, and then the other organs after insulin-containing blood returns from the heart. | Insulin is constantly released into the blood stream at a basal level. When blood glucose levels rise after a meal containing carbohydrates (sugars and starches), increased intracellular glucose in the beta cells triggers a series of processes that change the intracellular calcium concentration. First, the calcium concentration rises and stays elevated for 5-10 minutes, and then it spikes in intervals of roughly 3 minutes. Whenever calcium concentrations are high, insulin-containing vesicles merge with the plasma membrane, releasing insulin into the blood stream in the same temporal pattern as the calcium spikes<ref>DOI: 10.1007/s00125-009-1314-y</ref>. The insulin is released into the bloodstream, where it first reaches the liver via the portal vein<ref>"https://www.diabetesselfmanagement.com/diabetes-resources/definitions/portal-vein/"</ref>, and then the other organs after insulin-containing blood returns from the heart. | ||
| - | [[Image:Insulin receptor cartoon.gif|frame|Artist's conception of insulin receptor (purple) binding to insulin outside of the cell and being phosphorylated inside of the cell]]Liver cells and other cells contain [[insulin receptor|receptors]] with insulin binding sites on the cell surface. These intermembrane receptors<ref>https://pdb101.rcsb.org/motm/182</ref> change conformation upon insulin binding, activating intracellular tyrosine kinase domains<ref name="gutmann">DOI: 10.1083/jcb.201711047</ref>. This starts a signalling cascade within the cell that changes glucose uptake as well as carbohydrate and lipid metabolism. The liver stores available glucose in the form of glycogen and, when there is additional glucose, converts it into fatty acids and carbon dioxide. Cells in other organs (e.g. adipose tissue, muscles) also alter their metabolism depending on insulin levels. | + | [[Image:Insulin receptor cartoon.gif|frame|Artist's conception of insulin receptor (purple) binding to insulin outside of the cell and being phosphorylated inside of the cell]]Liver cells and other cells contain [[insulin receptor|receptors]] with insulin binding sites on the cell surface. These intermembrane receptors<ref>https://pdb101.rcsb.org/motm/182</ref> change conformation upon insulin binding, activating intracellular tyrosine kinase domains<ref name="gutmann">DOI: 10.1083/jcb.201711047</ref>. This starts a signalling cascade within the cell that changes glucose uptake as well as carbohydrate and lipid metabolism<ref>http://www.vivo.colostate.edu/hbooks/pathphys/endocrine/pancreas/insulin_phys.html</ref>. The liver stores available glucose in the form of glycogen and, when there is additional glucose, converts it into fatty acids and carbon dioxide. Cells in other organs (e.g. adipose tissue, muscles) also alter their metabolism depending on insulin levels. |
Once insulin binds to its receptor, the receptor is removed from the plasma membrane, insulin is degraded and the receptor is recycled<ref>PMID:28974775</ref>. The majority of insulin is removed from the bloodstream this way on its first pass through the liver. The half-life of insulin in the blood plasma is about about 4-6 minutes<ref>DOI:10.1210/edrv.19.5.0349</ref>. | Once insulin binds to its receptor, the receptor is removed from the plasma membrane, insulin is degraded and the receptor is recycled<ref>PMID:28974775</ref>. The majority of insulin is removed from the bloodstream this way on its first pass through the liver. The half-life of insulin in the blood plasma is about about 4-6 minutes<ref>DOI:10.1210/edrv.19.5.0349</ref>. | ||
| Line 48: | Line 53: | ||
<StructureSection load='' size='350' side='right' scene='82/821037/Ribbon/1' caption=''> | <StructureSection load='' size='350' side='right' scene='82/821037/Ribbon/1' caption=''> | ||
===Structure of mature insulin monomer=== | ===Structure of mature insulin monomer=== | ||
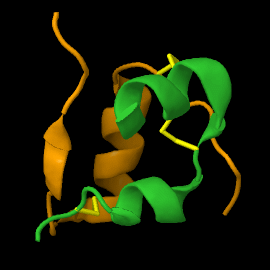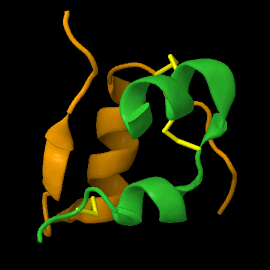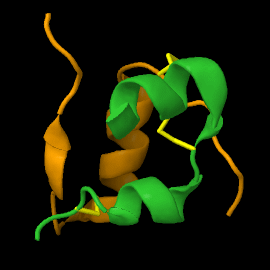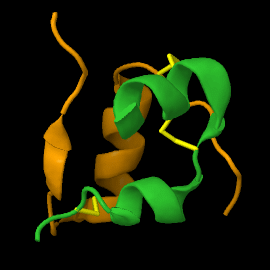
| - | <scene name='82/821037/Ribbon/1'> | + | Mature insulin (<scene name='82/821037/Ribbon/1'>reload initial figure</scene>) contains two chains, A <jmol> |
| + | <jmolLink> | ||
| + | <script> select backbone and *:A; selectionHalos ON; delay 0.5;selectionHalos OFF; </script> | ||
| + | <text>(☼)</text> | ||
| + | </jmolLink> | ||
| + | </jmol> and B <jmol> | ||
| + | <jmolLink> | ||
| + | <script> select backbone and *:B; selectionHalos ON; delay 0.5;selectionHalos OFF; </script> | ||
| + | <text>(☼)</text> | ||
| + | </jmolLink> | ||
| + | </jmol>, held together by disulfide bonds <jmol> | ||
| + | <jmolLink> | ||
| + | <script> select (*:A or *:B) and cys and (*.CA or *.SG); selectionHalos ON; delay 0.5;selectionHalos OFF; </script> | ||
| + | <text>(☼)</text> | ||
| + | </jmolLink> | ||
| + | </jmol> and non-covalent interactions. This structure, determined by Dorothy Hodgkin in 1969 using X-ray crystallography, was one of the [[Highest_impact_structures|first protein structures]] to be solved. The <scene name='82/821037/Spacefilling/2'>surface of insulin</scene> contains quite a few hydrophobic side chains, which form protein:protein contacts when insulin forms hexamers or binds to its receptor. Select coloring of the side chains below to explore the surface properties. | ||
Sidechains colored by <jmol> | Sidechains colored by <jmol> | ||
| Line 73: | Line 93: | ||
[[Image:Proinsulin.jpg|400 px]] | [[Image:Proinsulin.jpg|400 px]] | ||
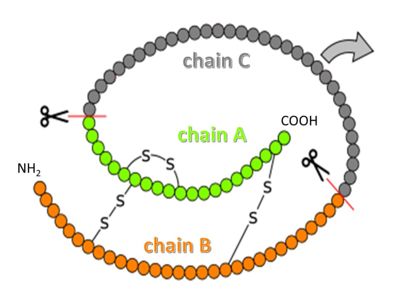
| - | Insulin | + | Insulin is synthesized as so-called preproinsulin and gets targeted into the ER and Golgi via a signal sequence, which then is removed to yield proinsulin. <scene name='82/821037/Proinsulin/1'>Proinsulin</scene> is processed by several proteases in the Golgi apparatus to form 3 separate chains, named B, C and A (see figure above)<ref>PMID:15289650</ref>. Chains B and A are linked through disulfide bonds, and are the components of mature insulin. |
<jmol> | <jmol> | ||
| Line 94: | Line 114: | ||
<jmolLink> | <jmolLink> | ||
<script>anim mode loop; anim on</script> | <script>anim mode loop; anim on</script> | ||
| - | <text>disordered</text></jmolLink></jmol> in the proinsulin structure, is no longer connected to insulin after processing | + | <text>disordered</text></jmolLink></jmol> in the proinsulin structure, is no longer connected to insulin after processing, but is secreted into the blood stream as so-called C-peptide. C-peptide has a role in diagnostics; it is used as a measure of how much insulin is made endogenously in patients who receive insulin as treatment (insulin is administered as mature protein, so it does not contain any C-peptide)<ref>https://en.wikipedia.org/wiki/C-peptide</ref>. |
===Storage=== | ===Storage=== | ||
| - | Before insulin is secreted, it is able to pair-up with itself and form a dimer by forming hydrogen bonds between the ends of two B-chains, which then combine in threes to form a hexamer (a trimer of dimers to be exact). In crystal structures, insulin occurs in two states, <scene name='82/821037/Rvst/1'>T or R</scene>. In an <scene name='82/821037/Rvst/2'>animation</scene> (takes long to load), you can see the conformational change in chain B while maintaining the | + | Before insulin is secreted, it is able to pair-up with itself and form a dimer by forming hydrogen bonds between the ends of two B-chains, which then combine in threes to form a hexamer (a trimer of dimers to be exact). In crystal structures, insulin occurs in two states, <scene name='82/821037/Rvst/1'>T or R</scene>. In an <scene name='82/821037/Rvst/2'>animation</scene> (takes long to load), you can see the conformational change in chain B while maintaining the organization of the dimer. |
(A) <scene name='82/821037/T6/1'>T6 hexamer (e.g. structure 4INS) </scene>, (B) T3Rf3 hexamer (e.g. 1TRZ) and (C) R6 hexamer (e.g. 1ZNJ). Small changes in the sequence of insulin change how fast hexamers fall apart into monomers. This is used to make insulin preparations that give off low levels of insulin for a longer time, or high levels of insulin for a short time when injected as microcrystals in depots under the skin. | (A) <scene name='82/821037/T6/1'>T6 hexamer (e.g. structure 4INS) </scene>, (B) T3Rf3 hexamer (e.g. 1TRZ) and (C) R6 hexamer (e.g. 1ZNJ). Small changes in the sequence of insulin change how fast hexamers fall apart into monomers. This is used to make insulin preparations that give off low levels of insulin for a longer time, or high levels of insulin for a short time when injected as microcrystals in depots under the skin. | ||
===Receptor interaction=== | ===Receptor interaction=== | ||
| - | The insulin receptor belongs to the class of tyrosine kinase receptors. Many of these receptors occur as monomers that dimerize upon ligand binding, bringing the intracellular tytrosine kinase domains (the endodomains) into close vicinity. In contrast, the insulin receptor (just like the closely related IGF-1 receptor) is a dimer even in the absence of ligand, crosslinked by disulfide bridges. The unliganded receptor ectodomain has the shape of a Λ (an inverted V), keeping the transmembrane segments and the endodomains at a distance. | + | The insulin receptor belongs to the class of [[tyrosine kinase]] receptors. Many of these receptors occur as monomers that dimerize upon ligand binding, bringing the intracellular tytrosine kinase domains (the endodomains) into close vicinity. In contrast, the insulin receptor (just like the closely related [[IGF1#Stimulating_interaction_:_IGF-1_-_IGF-1R|IGF-1 receptor]]) is a dimer even in the absence of ligand, crosslinked by disulfide bridges. The unliganded receptor ectodomain has the shape of a Λ (an inverted V), keeping the transmembrane segments and the endodomains at a distance. |
| - | Cryo-electronmicroscopy studies have shown a [http://proteopedia.org/wiki/images/0/05/LambdaTee.gif Λ to T transition] when insulin binds. One structure resolves four <scene name='82/821037/Spacefilling/5'>insulin binding sites</scene> (1, 1', 2, 2') per receptor dimer<ref name="ecto">DOI:10.1101/679233 </ref> (coordinates not yet available). Contacts with insulin are distinct in site 1 vs. site 2, as are the conformations of insulin. For comparison to the initial scene, here is another view of the <scene name='82/821037/Ribbon/2'>contact residues</scene>. | + | [[Cryo-EM|Cryo-electronmicroscopy]] studies have shown a [http://proteopedia.org/wiki/images/0/05/LambdaTee.gif Λ to T transition] when insulin binds. One structure resolves four <scene name='82/821037/Spacefilling/5'>insulin binding sites</scene> (1, 1', 2, 2') per receptor dimer<ref name="ecto">DOI:10.1101/679233 </ref> (coordinates not yet available). Contacts with insulin are distinct in site 1 vs. site 2, as are the conformations of insulin<ref>PMID:25092300</ref>. For comparison to the initial scene, here is another view of the <scene name='82/821037/Ribbon/2'>contact residues</scene>. |
| - | <scene name='82/821037/Receptor_bound/1'>Binding sites 1 and 1'</scene> had already been characterized in previous structures<ref>PMID:29512653</ref><ref name="gutmann"/>. Insulin makes contacts with both the N-terminal domain and the CT alpha helix, structural elements that are far apart in the unliganded conformation. Thus, binding of insulin in site 1 requires a large structural rearrangement that activates the tyrosine kinase activity of the endodomain by bringing the two kinase domains close for autophosphorylation. | + | <scene name='82/821037/Receptor_bound/1'>Binding sites 1 and 1'</scene> had already been characterized in previous structures<ref>PMID:29512653</ref><ref name="gutmann"/>. Insulin makes contacts with both the N-terminal domain <jmol> |
| + | <jmolLink> | ||
| + | <script> select *:A; selectionHalos ON; delay 0.5;selectionHalos OFF; </script> | ||
| + | <text>(☼)</text> | ||
| + | </jmolLink> | ||
| + | </jmol> and the CT alpha helix <jmol> | ||
| + | <jmolLink> | ||
| + | <script> select *:P; selectionHalos ON; delay 0.5;selectionHalos OFF; </script> | ||
| + | <text>(☼)</text> | ||
| + | </jmolLink> | ||
| + | </jmol>, structural elements that are far apart in the unliganded conformation. Thus, binding of insulin in site 1 requires a large structural rearrangement that activates the tyrosine kinase activity of the endodomain by bringing the two kinase domains close for autophosphorylation. | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | ==Insulin 3D structures== | ||
| + | [[Insulin 3D Structures]] | ||
==See also== | ==See also== | ||
| Line 118: | Line 151: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
Insulin is a peptide hormone that helps to maintain blood sugar within a healthy range by regulating carbohydrate and lipid metabolism throughout the body. It is secreted by specialized cells in the pancreas and acts by binding to insulin receptors on other cells. Insulin in its mature form contains two peptide chains connected by disulfide crosslinks, and occurs either as monomer or as hexamer. Administering insulin in carefully determined doses at the appropriate times is used in managing diabetes, a chronic condition where the body fails to maintain blood sugar levels by itself.
- Proinsulin is the insulin precursor which is converted in the pancreas to insulin and C-peptide[1].
- Leginsulin is an insulin-like peptide found in legumes [2].
- Insulin lispro is recombinant insulin which has more rapid absorption than regular insulin[3]. See also Insulin lispro.
- Insulin glargine is a long-acting insulin [4]. See also Insulin glargine.
Contents |
Other Proteopedia pages about or relating to insulin
This page is for an audience with some basic knowledge of biochemistry. If you encounter terms you don't know, such as disulfide bond or hexamer, you are encouraged to look them up on Proteopedia or elsewhere. Sometimes, links are already provided, and sometimes a close look at the 3D figures will already give you an idea. Below are listed other pages on the topic, some more basic and some more detailed.
Diabetes & Hypoglycemia
Insulin (Hebrew)
Insulin aspart
Insulin and pro-insulin (Hebrew)
Insulin-Degrading Enzyme
Molecular_Playground/Insulin
Insulin Structure & Function
Insulin signal transduction pathway
Proteins that interact with insulin
Insulin Receptor UMAss
Insulin Receptor - kinase_domain (Hebrew)
Insulin receptor
Insulin-like growth factor receptor
Tutorials we would love to see
Tutorial: what molecules play a role in diabetes? (TBD)
Tutorial: how does insulin regulate my blood sugar? (TBD)
Function
Our blood sugar level (i.e. glucose concentration) varies over time with food intake and exercise, but has to remain in a narrow range so we stay healthy (not become hyperglycemic or hypoglycemic). Insulin, together with glucagon, regulates blood sugar levels by changing fuel metabolism in all metabolic cells (e.g. liver, fat, kidney, muscle and nerve cells) on a timescale of minutes and hours[5][6][7]. In simplest terms, the presence of insulin in the blood signals the well-fed stage, while the presence of glucagon signals the fasting stage.
Biosynthesis and processing of insulin occurs in the beta cells of the pancreas. The beta cells are found in the islets of Langerhans, which also contain the alpha cells that synthesize glucagon. Insulin is made as a 110 amino acid pre-proinsulin, which is processed to the mature 51 amino acid insulin and targeted to secretory vesicles. In a healthy adult, about 200 units of insulin are available in the pancreas, of which 30-50 units are secreted daily[8]. With a unit of insulin corresponding to 0.0347 mg mature insulin [9], the body contains only about 7 mg of insulin. In its stored form, insulin is a hexamer complexed to zinc ions.
Insulin is constantly released into the blood stream at a basal level. When blood glucose levels rise after a meal containing carbohydrates (sugars and starches), increased intracellular glucose in the beta cells triggers a series of processes that change the intracellular calcium concentration. First, the calcium concentration rises and stays elevated for 5-10 minutes, and then it spikes in intervals of roughly 3 minutes. Whenever calcium concentrations are high, insulin-containing vesicles merge with the plasma membrane, releasing insulin into the blood stream in the same temporal pattern as the calcium spikes[10]. The insulin is released into the bloodstream, where it first reaches the liver via the portal vein[11], and then the other organs after insulin-containing blood returns from the heart.
Once insulin binds to its receptor, the receptor is removed from the plasma membrane, insulin is degraded and the receptor is recycled[15]. The majority of insulin is removed from the bloodstream this way on its first pass through the liver. The half-life of insulin in the blood plasma is about about 4-6 minutes[16].
Disease and Treatment
In patients with diabetes, insulin signalling is compromised[17]. In type I diabetes, insulin is not produced sufficiently because the beta cells in the pancreas are absent or compromised by an autoimmune event. The onset of type 1 diabetes is rapid, and often occurs early in life ("juvenile diabetes"). In type II diabetes, insulin is secreted at normal or even elevated levels, but the target cells do not respond properly, for reasons not yet understood[18]. The onset of type 2 diabetes is gradual (over years), and there are warning signs (like insulin resistance) that in some cases helps to avert the onset by certain changes in lifestyle and diet[19].
Type 1 diabetes is managed by administering insulin. The challenge is to keep the blood sugar level in a healthy range while conditions change (food intake, exercise, illness). Insulin can not be given orally because it is broken down in the digestive tract. Instead, it is administered via subcutaneous depots or in other non-oral forms). There are long-acting and short-acting preparations of insulin to provide the basal level and to manage spikes in blood glucose levels, respectively. Type 2 diabetes is managed in a variety of ways, which may include administration of extra insulin to partially counteract the diminished signalling capacity.
Structure
| |||||||||||
Insulin 3D structures
See also
Insulin @RCSB
Insulin @Wikipedia
Insulin @PDBe
Diabetes @RCSB
References
- ↑ Kitabchi AE. Proinsulin and C-peptide: a review. Metabolism. 1977 May;26(5):547-87. PMID:403392 doi:10.1016/0026-0495(77)90099-3
- ↑ Hashidume T, Sakano T, Mochizuki A, Ito K, Ito S, Kawarasaki Y, Miyoshi N. Identification of soybean peptide leginsulin variants in different cultivars and their insulin-like activities. Sci Rep. 2018 Nov 15;8(1):16847. PMID:30442953 doi:10.1038/s41598-018-35331-5
- ↑ Wilde MI, McTavish D. Insulin lispro: a review of its pharmacological properties and therapeutic use in the management of diabetes mellitus. Drugs. 1997 Oct;54(4):597-614. PMID:9339963 doi:10.2165/00003495-199754040-00006
- ↑ Gillies PS, Figgitt DP, Lamb HM. Insulin glargine. Drugs. 2000 Feb;59(2):253-60; discussion 261-2. PMID:10730548 doi:10.2165/00003495-200059020-00009
- ↑ https://www.yourhormones.info/hormones/insulin/
- ↑ Sonksen P, Sonksen J. Insulin: understanding its action in health and disease. Br J Anaesth. 2000 Jul;85(1):69-79. PMID:10927996
- ↑ Weiss MA, Lawrence MC. A thing of beauty: Structure and function of insulin's "aromatic triplet". Diabetes Obes Metab. 2018 Sep;20 Suppl 2:51-63. doi: 10.1111/dom.13402. PMID:30230175 doi:http://dx.doi.org/10.1111/dom.13402
- ↑ https://www.britannica.com/science/insulin
- ↑ https://www.who.int/biologicals/expert_committee/BS_2143_Human_Recombinant_Insulin_final.pdf
- ↑ Henquin JC. Regulation of insulin secretion: a matter of phase control and amplitude modulation. Diabetologia. 2009 May;52(5):739-51. doi: 10.1007/s00125-009-1314-y. Epub 2009, Mar 14. PMID:19288076 doi:http://dx.doi.org/10.1007/s00125-009-1314-y
- ↑ "https://www.diabetesselfmanagement.com/diabetes-resources/definitions/portal-vein/"
- ↑ https://pdb101.rcsb.org/motm/182
- ↑ 13.0 13.1 Gutmann T, Kim KH, Grzybek M, Walz T, Coskun U. Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J Cell Biol. 2018 May 7;217(5):1643-1649. doi: 10.1083/jcb.201711047. Epub 2018, Feb 16. PMID:29453311 doi:http://dx.doi.org/10.1083/jcb.201711047
- ↑ http://www.vivo.colostate.edu/hbooks/pathphys/endocrine/pancreas/insulin_phys.html
- ↑ Haeusler RA, McGraw TE, Accili D. Biochemical and cellular properties of insulin receptor signalling. Nat Rev Mol Cell Biol. 2018 Jan;19(1):31-44. doi: 10.1038/nrm.2017.89. Epub 2017 , Oct 4. PMID:28974775 doi:http://dx.doi.org/10.1038/nrm.2017.89
- ↑ Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998 Oct;19(5):608-24. doi: 10.1210/edrv.19.5.0349. PMID:9793760 doi:http://dx.doi.org/10.1210/edrv.19.5.0349
- ↑ https://www.endotext.org/section/diabetes/
- ↑ Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J Clin Invest. 2016 Jan;126(1):12-22. doi: 10.1172/JCI77812. Epub 2016 Jan 4. PMID:26727229 doi:http://dx.doi.org/10.1172/JCI77812
- ↑ doi: https://dx.doi.org/10.1530/endoabs.56.PL5
- ↑ Davidson HW. (Pro)Insulin processing: a historical perspective. Cell Biochem Biophys. 2004;40(3 Suppl):143-58. PMID:15289650
- ↑ https://en.wikipedia.org/wiki/C-peptide
- ↑ doi: https://dx.doi.org/10.1101/679233
- ↑ Menting JG, Yang Y, Chan SJ, Phillips NB, Smith BJ, Whittaker J, Wickramasinghe NP, Whittaker LJ, Pandyarajan V, Wan ZL, Yadav SP, Carroll JM, Strokes N, Roberts CT Jr, Ismail-Beigi F, Milewski W, Steiner DF, Chauhan VS, Ward CW, Weiss MA, Lawrence MC. Protective hinge in insulin opens to enable its receptor engagement. Proc Natl Acad Sci U S A. 2014 Aug 4. pii: 201412897. PMID:25092300 doi:http://dx.doi.org/10.1073/pnas.1412897111
- ↑ Scapin G, Dandey VP, Zhang Z, Prosise W, Hruza A, Kelly T, Mayhood T, Strickland C, Potter CS, Carragher B. Structure of the Insulin Receptor-Insulin Complex by Single Particle CryoEM analysis. Nature. 2018 Feb 28. pii: nature26153. doi: 10.1038/nature26153. PMID:29512653 doi:http://dx.doi.org/10.1038/nature26153
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Alexander Berchansky, Jaime Prilusky, David Canner