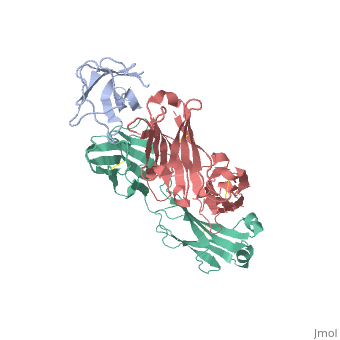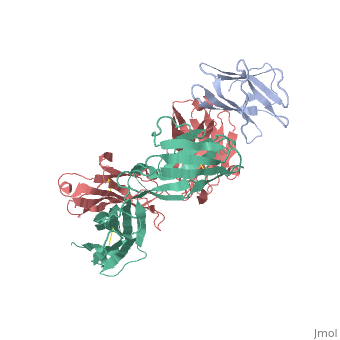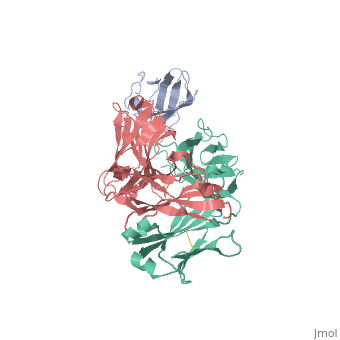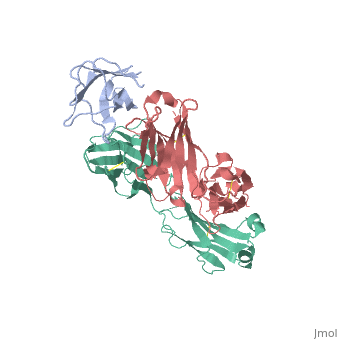
2vxq
From Proteopedia
(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:2vxq.png|left|200px]] | ||
| - | < | + | ==Crystal structure of the major grass pollen allergen Phl p 2 in complex with its specific IgE-Fab== |
| - | + | <StructureSection load='2vxq' size='340' side='right'caption='[[2vxq]], [[Resolution|resolution]] 1.90Å' scene=''> | |
| - | You may | + | == Structural highlights == |
| - | + | <table><tr><td colspan='2'>[[2vxq]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] and [https://en.wikipedia.org/wiki/Phleum_pratense Phleum pratense]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2VXQ OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2VXQ FirstGlance]. <br> | |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.9Å</td></tr> | |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2vxq FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2vxq OCA], [https://pdbe.org/2vxq PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2vxq RCSB], [https://www.ebi.ac.uk/pdbsum/2vxq PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2vxq ProSAT]</span></td></tr> | |
| - | + | </table> | |
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/MPAP2_PHLPR MPAP2_PHLPR] | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/vx/2vxq_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2vxq ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | We report the three-dimensional structure of the complex between the major respiratory grass pollen allergen Phl p 2 and its specific human IgE-derived Fab. The Phl p 2-specific human IgE Fab has been isolated from a combinatorial library constructed from lymphocytes of a pollen allergic patient. When the variable domains of the IgE Fab were grafted onto human IgG1, the resulting Ab (huMab2) inhibited strongly the binding of allergic patients' IgE to Phl p 2 as well as allergen-induced basophil degranulation. Analysis of the binding of the allergen to the Ab by surface plasmon resonance yielded a very low dissociation constant (K(D) = 1.1 x 10(-10) M), which is similar to that between IgE and Fcepsilon;RI. The structure of the Phl p 2/IgE Fab complex was determined by x-ray crystallography to 1.9 A resolution revealing a conformational epitope (876 A(2)) comprised of the planar surface of the four-stranded anti-parallel beta-sheet of Phl p 2. The IgE-defined dominant epitope is discontinuous and formed by 21 residues located mostly within the beta strands. Of the 21 residues, 9 interact directly with 5 of the 6 CDRs (L1, L3, H1, H2, H3) of the IgE Fab predominantly by hydrogen bonding and van der Waals interactions. Our results indicate that IgE Abs recognize conformational epitopes with high affinity and provide a structural basis for the highly efficient effector cell activation by allergen/IgE immune complexes. | ||
| - | + | High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography.,Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G, Vieths S, Lupinek C, Ebner C, Valenta R, Markovic-Housley Z J Immunol. 2009 Feb 15;182(4):2141-51. PMID:19201867<ref>PMID:19201867</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | <div class="pdbe-citations 2vxq" style="background-color:#fffaf0;"></div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
==See Also== | ==See Also== | ||
| - | *[[Monoclonal | + | *[[Monoclonal Antibodies 3D structures|Monoclonal Antibodies 3D structures]] |
| - | + | == References == | |
| - | == | + | <references/> |
| - | < | + | __TOC__ |
| + | </StructureSection> | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| + | [[Category: Large Structures]] | ||
[[Category: Phleum pratense]] | [[Category: Phleum pratense]] | ||
| - | [[Category: Ebner | + | [[Category: Ebner C]] |
| - | [[Category: Flicker | + | [[Category: Flicker S]] |
| - | [[Category: Lupinek | + | [[Category: Lupinek C]] |
| - | [[Category: Madritsch | + | [[Category: Madritsch C]] |
| - | [[Category: Markovic-Housley | + | [[Category: Markovic-Housley Z]] |
| - | [[Category: Padavattan | + | [[Category: Padavattan S]] |
| - | [[Category: Randow | + | [[Category: Randow S]] |
| - | [[Category: Reese | + | [[Category: Reese G]] |
| - | [[Category: Schirmer | + | [[Category: Schirmer T]] |
| - | [[Category: Valenta | + | [[Category: Valenta R]] |
| - | [[Category: Vieths | + | [[Category: Vieths S]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Crystal structure of the major grass pollen allergen Phl p 2 in complex with its specific IgE-Fab
| |||||||||||
Categories: Homo sapiens | Large Structures | Phleum pratense | Ebner C | Flicker S | Lupinek C | Madritsch C | Markovic-Housley Z | Padavattan S | Randow S | Reese G | Schirmer T | Valenta R | Vieths S