User:Cristiane Custodio Ross Matheus/Sandbox 1
From Proteopedia
< User:Cristiane Custodio Ross Matheus(Difference between revisions)
| (27 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='1ZB9' size='450' side='right | + | <StructureSection load='1ZB9' size='450' side='right' caption="'Organic Hydroperoxide Resistance Protein (PDB code [[1ZB9]])." > |
| - | ==Introduction== | + | == Introduction == |
| - | + | '''Ohr''' is a thiol dependent antioxidant enzyme that belongs to the Ohr/OsmC family, present in bacteria and fungi, and which has the function of protecting these microorganisms from oxidative stress. It is evident, therefore, that this protein plays a central role in bacterial defense against oxidants from the host's reaction to infection. With the progress of studies on this compound it was found that Ohr are the essential actors in the decomposition of hydroperoxides of fatty acids and peroxynitrite, that is, it has these compounds as its main substrate, not presenting much efficiency in the decomposition of other types of oxidants. Within this context it is important to point out that although similar to peroxiredoxins the Ohr are not seen as such. Peroxiredoxins are nothing more than proteins that also have antioxidant action but have a structure both primary and tertiary, quite different from Ohr enzymes.. Another important point to be raised is the relevance of ohr studies for the creation of new drugs, since it is an enzyme whose structure is characteristic only of bacteria and fungi, not presenting similar structure in animals and plants. Some examples of pathogenic bacteria expressing Ohr proteins are Pseudomonas aeruginosa, Vibrio cholerae and Xylella fastidiosa. It is important to note that this protein varies according to the microorganism studied, having differences in its regulation and expression, and the focus of this page is the Ohr protein of Xylella Fastidiosa. | |
| - | '''Ohr''' | + | |
| - | + | ||
==Organic Hydroperoxide Resistance Protein from Xylella fastidiosa (Ohr)== | ==Organic Hydroperoxide Resistance Protein from Xylella fastidiosa (Ohr)== | ||
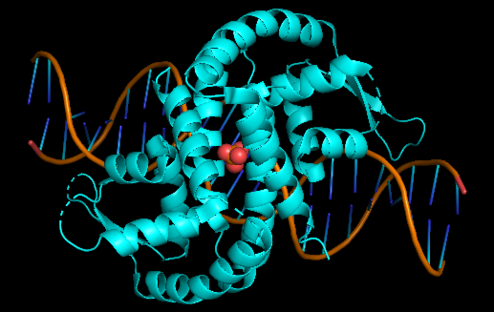
| - | + | '''Xylella fastidiosa''' is a gram - negative bacterium, restricted colonizer of plant xylem, known to cause diseases in several monocotyledons and dicotyledons of great economic importance. Addressing the pathogen-host relationship, it was possible to observe that the plant releases reactive oxygen species that function as microbicide agents. Among the ROS produced are fatty acid hydroperoxides, generated from an oxidation reaction catalyzed by lipoxygenase enzymes. To defend itself from this oxidative attack Xylella produces the Enzyme Ohr. Within this topic it was observed that in a situation of increased stress there is no increase in the amount of enzyme, concluding that in this bacterium there is no regulatory mechanism as for most OM so that have the Ohr gene | |
| - | '''Xylella fastidiosa''' | + | [[Image:OHRR_(2).png|left|494px]]<br /> |
| - | + | ||
==Ohr gene regulation== | ==Ohr gene regulation== | ||
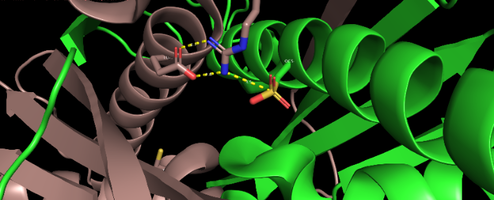
| + | '''OhrR''' is a transcriptional factor characterized by being a repressive protein and the main regulatory factor of the Ohr gene in most microorganisms. This protein is found on the promoter of the Ohr gene and has its structure altered when oxidized when it comes into contact with hydroperoxides of fatty acids and peroxynitrites. With the change of conformation generated by the oxidation of OhrR occurs its detachment from the promoter of the gene, making it more accessible to RNA polymerase, resulting in an overexpression of the Ohr gene. In addition to OhrR were also found in some microorganisms different regulatory means for the Ohr gene. An example is positive regulation by the alternative sigma factor, present in some microorganisms | ||
| + | [[Image:Screenshot 20211212-110409~2.png|right|494px]]<br /> | ||
| + | ==Structural features== | ||
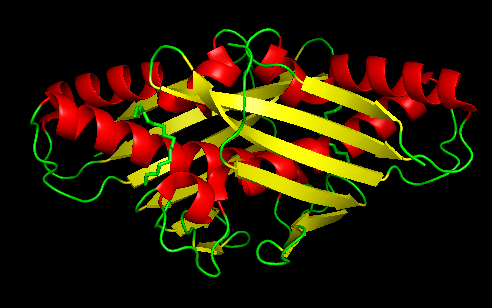
| - | + | Ohr presents distinct structural and biochemical characteristics when compared to cys-based mammalian peroxidases. This protein has a barrel-like structure formed by a tightly folded homodhermer, in which two β sheets of six ribbons involve two αhelix. There are two active sites in the enzyme that are located at the dye interface on opposite sides of the protein.The architecture of the Ohr catalytic site is composed of two cysteines, peroxidatic and resolution. Peroxidatic cysteine, located in one of α central helixs, has as function the direct reaction with hydroperoxides, forming a sulphenic acid. This in turn condenses with resolution cysteine to form an intramoleculardisulfide bond. In addition to the two cys amino acids there is also the presence of a catalytic arginine and a glutamate that has extreme importance in the activity of the enzyme Ohr, since they help stabilize the Cys in its thiolato state through polar interactions, increasing its nucleophilicity.The carboxylic group of catalytic glutamate guides the Guanidine group of Arginine towards Cysteine, in a configuration that seems to be ideal for the reduction of organic hydroperoxides. This described stabilization process is the so-called closed state of the catalytic triad. In the open state, there is a disruption of interactions and consequent conformational change that ends up exposing the residue to the solvent, the latter being the most conducive to the reduction of Ohr | |
| + | [[Image:OHR.png|center|494px]]<br /> | ||
| + | ==3D structures of Ohr== | ||
| + | [[Organic hydroperoxide resistance protein]] | ||
| - | + | </StructureSection> | |
| - | ==Structural features== | ||
| - | The resulting model 2pex shows the biologically relevant dimer of the protein. Each subunit of the dimer is composed of six α-helices and 3 β-strands, as indicated below (left). The specific residues corresponding to these regions of secondary structure are as follows: α1 (residues 21–39), α2 (residues 47–58), β1(residues 62–63), α3 (residues 64–71), α4 (residues 75–87), β2 (residues 91–94), β3 (residues 104–107), α5 (residues 109–129), α6 (residues 133–151), and three-ten helices 1a (residues 13–15) and 1b (residues 17–19). The longest α-helix, α5, has a notable kink at residue G119. The dimerization interface is largely formed by three-ten helices (1a, 1b) and α1, α5, and α6 from each subunit. The extensive dimerization domain buries 5391Ų. | ||
| - | + | ==References== | |
| - | + | *ALEGRIA, Thiago Geronimo Pires. Caracterização cinética e busca de inibidores de Ohr (Organic Hydroperoxide Resistance protein) de Xylella fastidiosa. 2012. 117 f. Tese (Doutorado) - Curso de Biologia, Universidade de São Paulo, São Paulo, 2012. Disponível em: https://teses.usp.br/teses/disponiveis/41/41131/tde-23072012-160418/publico/ThiagoGeronimo_Alegria.pdf. Acesso em: 13 nov. 2021. | |
| - | + | *CUSSIOL, Jose Renato Rosa. Caracterização funcional de uma nova proteína antioxidante: Ohr (Organic Hidroperoxide Resistance Protein). Vias de redução e expressão em Xylella fastidiosa. 2010. 218 f. Tese (Doutorado) - Curso de Biociências, Departamento de Biologia Evolutiva e Genética, Usp, São Paulo, 2010. Disponível em: https://www.teses.usp.br/teses/disponiveis/41/41131/tde-21072010-161740/publico/Jose_Renato_Cussiol_versao_completa.pdf. Acesso em: 13 nov. 2021. | |
| - | + | *NETTO, Luis.Structural insights on the efficient catalysis of hydroperoxide reduction by Ohr: Crystallographic and molecular dynamics approaches. PLOS one, São Paulo, v. -, n.-,p.1-23,maio 2018. | |
| - | + | *NETTO, Luis. Structural Switches along Organic Hydroperoxide Resistance Protein Catalytic Cycle. Acs Catalysis, São Paulo, v. -, n. -, p. 6587-6602, maio 2020. | |
Current revision
| |||||||||||
References
- ALEGRIA, Thiago Geronimo Pires. Caracterização cinética e busca de inibidores de Ohr (Organic Hydroperoxide Resistance protein) de Xylella fastidiosa. 2012. 117 f. Tese (Doutorado) - Curso de Biologia, Universidade de São Paulo, São Paulo, 2012. Disponível em: https://teses.usp.br/teses/disponiveis/41/41131/tde-23072012-160418/publico/ThiagoGeronimo_Alegria.pdf. Acesso em: 13 nov. 2021.
- CUSSIOL, Jose Renato Rosa. Caracterização funcional de uma nova proteína antioxidante: Ohr (Organic Hidroperoxide Resistance Protein). Vias de redução e expressão em Xylella fastidiosa. 2010. 218 f. Tese (Doutorado) - Curso de Biociências, Departamento de Biologia Evolutiva e Genética, Usp, São Paulo, 2010. Disponível em: https://www.teses.usp.br/teses/disponiveis/41/41131/tde-21072010-161740/publico/Jose_Renato_Cussiol_versao_completa.pdf. Acesso em: 13 nov. 2021.
- NETTO, Luis.Structural insights on the efficient catalysis of hydroperoxide reduction by Ohr: Crystallographic and molecular dynamics approaches. PLOS one, São Paulo, v. -, n.-,p.1-23,maio 2018.
- NETTO, Luis. Structural Switches along Organic Hydroperoxide Resistance Protein Catalytic Cycle. Acs Catalysis, São Paulo, v. -, n. -, p. 6587-6602, maio 2020.