Journal:Acta Cryst D:S2059798320008116
From Proteopedia
(Difference between revisions)

| (24 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene=' | + | <StructureSection load='' size='450' side='right' scene='85/857784/Cv/1' caption=''> |
===Novel structure of the N-terminal helical domain of BibA, a Group B Streptococcus immunogenic bacterial adhesin=== | ===Novel structure of the N-terminal helical domain of BibA, a Group B Streptococcus immunogenic bacterial adhesin=== | ||
<big>Kartik Manne, Debasish Chattopadhyay, Vaibhav Agarwal, Anna M. Blom, Baldeep Khare, Srinivas Chakravarthy, Chungyu Chang, Hung Ton-That and Sthanam V. L. Narayana | <big>Kartik Manne, Debasish Chattopadhyay, Vaibhav Agarwal, Anna M. Blom, Baldeep Khare, Srinivas Chakravarthy, Chungyu Chang, Hung Ton-That and Sthanam V. L. Narayana | ||
| Line 5: | Line 5: | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| + | This publication describes a high–resolution X-ray crystallographic structure of the Group B Streptococcus BibA N-terminal fragment (BibA126-398), and a low-resolution structure of the full-length N-terminal domain (BibA34-400) determined using small angle X-ray scattering (SAXS). The association of the BibA N-terminal domain was localized to the C4BP α-chain. | ||
| - | <b>References</b><br> | + | <scene name='85/857784/Cv/5'>Cartoon representation of the BibA126–398 crystal structure</scene> shown as a rainbow model with color changing gradually from the N-terminus (blue) to the C-terminus (red) (PDB entry [[6poo]]). The antiparallel three-helix-bundle-motif repeats are labeled from the N-terminus to the C-terminus as MR1–MR4, respectively. The overall structure of BibA126–398 has an ~150 Å-long distorted rod shape composed of nine α-helices, a 310-helix and small loops connecting the helices. Interestingly, the nine α-helices adopt a unique pattern to form four antiparallel three-helix-bundle-motif repeats labeled <scene name='85/857784/Cv/7'>MR1</scene> (Asp126–Ser182), <scene name='85/857784/Cv/8'>MR2</scene> (Thr183–Lys235), <scene name='85/857784/Cv/9'>MR3</scene> (Glu236–Leu303) and <scene name='85/857784/Cv/10'>MR4</scene> (Gln304–Asp398). Topologically, the <scene name='85/857784/Cv/7'>N-terminus</scene> first assembles into an 18-residue α-helix (α1; Asp126–Ser142) and a small loop (L1; Leu143–Ser147), followed by the second α-helix (α2; Ser148–Ser160), which runs antiparallel to helix α1. Loop 2 (L2; Ser161–Asp163) proceeds α2, allowing the third α-helix (α3; Ser164–Leu195) to run antiparallel to α2. Among the first three helices, <scene name='85/857784/Cv/11'>α3 is more extended</scene>, comprising of 33 residues, of which residues Ser164–Ser182 are involved in forming the MR1 motif, while residues Thr183–Leu195 extend into the MR2 motif. This shared-helix pattern to form the motif is repeated to generate motifs <scene name='85/857784/Cv/12'>MR3 and MR4</scene>, which involve the remaining α-helices (α4–α9) and loops (L3–L8). The <scene name='85/857784/Cv/13'>C-terminal MR4 motif</scene> is composed of a longer stretch of residues (94) compared with MR1 (57 residues), MR2 (53 residues) and MR3 (67 residues). The α7 helix that is shared by MR3 and MR4 is the longest, with 54 residues (Ile281–Lys331). The longer MR4 motif harbors an additional 310-helix (Lys333–Ile335) between the α7 helix and loop 7 (L7), and an extended loop (L8, residues Leu357–Trp365) that is distinct from the other motifs. <scene name='85/857784/Cv/15'>Helices α3, α5 and α7</scene> bridge the three-helix-bundle motifs together to form an extended rod with a curved shape. |
| + | |||
| + | |||
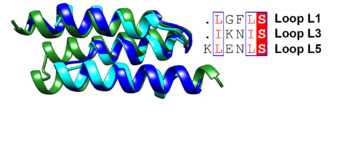
| + | [[Image:Figqqq.png|thumb|350px|left|Superposition of the antiparallel three-helix-bundle-motif repeats MR1 (dark blue), MR2 (cyan) and MR3 (forest green). Similar residues such as Leu and Ile are boxed and shown in a red font and the conserved serine residue is highlighted in red.]] | ||
| + | {{Clear}} | ||
| + | |||
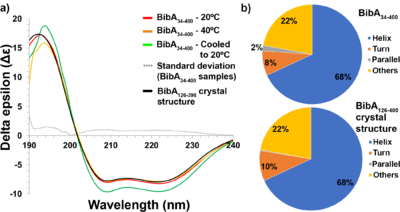
| + | [[Image:Fig-4-CD-final-edit7.png|thumb|400px|left|(a) Circular dichroism spectra of freshly prepared and intact BibA34–400 sample at 20°C (red), at 40°C (orange) and denatured at 80°C followed by cooling to 20°C (green), the standard deviation error between the BibA34–400 samples (gray dotted line) and the spectrum generated from the BibA126–398 crystal structure (black). Two negative peaks at 208 and 222 nm typical of a α-helical secondary structure were observed for the BibA34–400 sample. (b) Estimated secondary-structure content (%) of BibA34–400 sample in solution (top) and the spectrum generated from the BibA126–398 crystal structure (bottom).]] | ||
| + | {{Clear}} | ||
| + | |||
| + | [[Image:Fig5a-c-relabel-motifs-photoshop-edit.png|thumb|500px|left|(a) Pair distance distribution [P(r)] function of intact BibA34–400 protein. (b) Comparison of the experimental scattering profile (in blue) for BibA34–400 with profiles from a theoretical model (FoXS; green) derived from the proposed BibA34–400 model. (c) Fit of the crystal structure of BibA126–398 (cyan) and the proposed BibA34–400 (magenta) into the ab initio model of BibA34–400 calculated with ''DAMMIF''.]] | ||
| + | {{Clear}} | ||
| + | |||
| + | '''PDB reference:''' Novel structure of the N-terminal helical domain of BibA, a group B streptococcus immunogenic bacterial adhesin [[6poo]]. | ||
| + | <br><b>References</b><br> | ||
<references/> | <references/> | ||
</StructureSection> | </StructureSection> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.
![(a) Pair distance distribution [P(r)] function of intact BibA34–400 protein. (b) Comparison of the experimental scattering profile (in blue) for BibA34–400 with profiles from a theoretical model (FoXS; green) derived from the proposed BibA34–400 model. (c) Fit of the crystal structure of BibA126–398 (cyan) and the proposed BibA34–400 (magenta) into the ab initio model of BibA34–400 calculated with DAMMIF.](/wiki/images/thumb/9/94/Fig5a-c-relabel-motifs-photoshop-edit.png/500px-Fig5a-c-relabel-motifs-photoshop-edit.png)
