Copper Amine Oxidase
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
[[Image:TTS.png|thumb|left|alt=3-((3E)-4-HYDROXY-3-{[2-(4-HYDROXYPHENYL)ETHYL]IMINO}-6-OXOCYCLOHEXA-1,4-DIEN-1-YL)ALANINE.|Residue 382 is a tyrosine residue, modified into topa-quinone, shown here bound to tyramine.]] | [[Image:TTS.png|thumb|left|alt=3-((3E)-4-HYDROXY-3-{[2-(4-HYDROXYPHENYL)ETHYL]IMINO}-6-OXOCYCLOHEXA-1,4-DIEN-1-YL)ALANINE.|Residue 382 is a tyrosine residue, modified into topa-quinone, shown here bound to tyramine.]] | ||
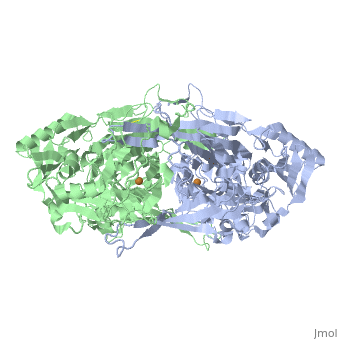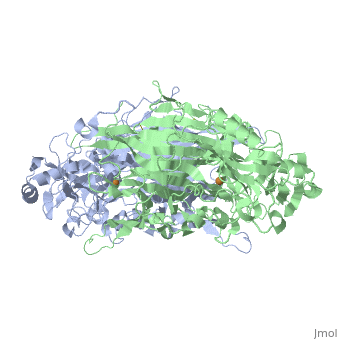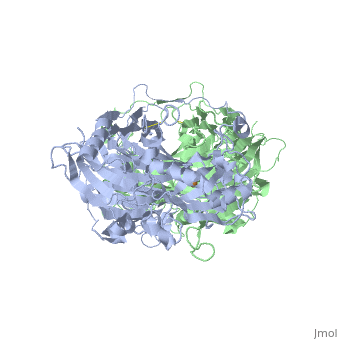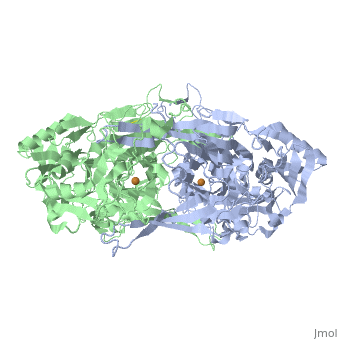
| - | '''Copper amine oxidases''' (CuAO) are classified as EC 1.4.3.6 in the EC number classification system of enzymes, and they belong to the larger class of [http://en.wikipedia.org/wiki/Oxidoreductase oxidoreductases]. [[2d1w]] is the tyramine-bound substrate Schiff-base intermediate of [http://en.wikipedia.org/wiki/Amine_oxidase_%28copper-containing%29 copper amine oxidase] derived from [http://en.wikipedia.org/wiki/Arthrobacter_globiformis Arthrobacter globiformis]. The structure of this enzyme was determined by Murakawa ''et al.'' in 2005, by x-ray diffraction<ref name="Murakawa">PMID:16487484</ref>. It consists of a disulfide-linked homodimer, with each subunit containing 638 residues. Each subunit also contains a <scene name='Sandbox_Reserved_331/Copper_ligand/5'>copper ligand</scene> near the active site, which is coordinated by three histidine residues. Located near the Cu<sup>2+</sup> ligand is a tyrosine residue that has been modified into topa-quinone ('''TPQ'''), which is also a cofactor in all other copper amine oxidases<ref name="Parsons">PMID:8591028</ref>. The active site of the enzyme is located near the center of the homodimer, which is connected to the outside of the enzyme by an extensively hydrated channel. It is suspected that the water helps to carry O<sub>2</sub> to the active site, as well as being used in the reaction itself<ref name="Mure">PMID:12135347</ref>. | + | '''Copper amine oxidases''' or '''primary amine oxidase''' (CuAO) are classified as EC 1.4.3.6 in the EC number classification system of enzymes, and they belong to the larger class of [http://en.wikipedia.org/wiki/Oxidoreductase oxidoreductases]. [[2d1w]] is the tyramine-bound substrate Schiff-base intermediate of [http://en.wikipedia.org/wiki/Amine_oxidase_%28copper-containing%29 copper amine oxidase] derived from [http://en.wikipedia.org/wiki/Arthrobacter_globiformis Arthrobacter globiformis]. The structure of this enzyme was determined by Murakawa ''et al.'' in 2005, by x-ray diffraction<ref name="Murakawa">PMID:16487484</ref>. It consists of a disulfide-linked homodimer, with each subunit containing 638 residues. Each subunit also contains a <scene name='Sandbox_Reserved_331/Copper_ligand/5'>copper ligand</scene> near the active site, which is coordinated by three histidine residues. Located near the Cu<sup>2+</sup> ligand is a tyrosine residue that has been modified into topa-quinone ('''TPQ'''), which is also a cofactor in all other copper amine oxidases<ref name="Parsons">PMID:8591028</ref>. The active site of the enzyme is located near the center of the homodimer, which is connected to the outside of the enzyme by an extensively hydrated channel. It is suspected that the water helps to carry O<sub>2</sub> to the active site, as well as being used in the reaction itself<ref name="Mure">PMID:12135347</ref>. <scene name='43/433611/Cv/1'>Cu coordination site</scene>. Water molecules are shown as red spheres. |
<div style="clear:left;"></div> | <div style="clear:left;"></div> | ||
| - | CuAO from ''Arthrobacter globiformis'' | + | CuAO from ''Arthrobacter globiformis'' is called '''phenylethylamine oxidase''' undergoes reduction of phenylethylamine and forms phenylacetaldehyde. |
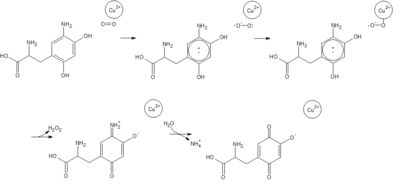
== Reaction Mechanism == | == Reaction Mechanism == | ||
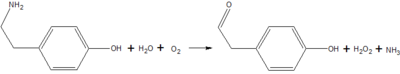
Copper amine oxidase catalyzes the oxidation of a primary amine to the corresponding aldehyde, yielding hydrogen peroxide and free ammonia. An example of this is the oxidation of [http://en.wikipedia.org/wiki/Tyramine tyramine]: | Copper amine oxidase catalyzes the oxidation of a primary amine to the corresponding aldehyde, yielding hydrogen peroxide and free ammonia. An example of this is the oxidation of [http://en.wikipedia.org/wiki/Tyramine tyramine]: | ||
Current revision
| |||||||||||
Additional Resources
References
- ↑ 1.0 1.1 Murakawa T, Okajima T, Kuroda S, Nakamoto T, Taki M, Yamamoto Y, Hayashi H, Tanizawa K. Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction. Biochem Biophys Res Commun. 2006 Apr 7;342(2):414-23. Epub 2006 Feb 8. PMID:16487484 doi:10.1016/j.bbrc.2006.01.150
- ↑ Parsons MR, Convery MA, Wilmot CM, Yadav KD, Blakeley V, Corner AS, Phillips SE, McPherson MJ, Knowles PF. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution. Structure. 1995 Nov 15;3(11):1171-84. PMID:8591028
- ↑ Mure M, Mills SA, Klinman JP. Catalytic mechanism of the topa quinone containing copper amine oxidases. Biochemistry. 2002 Jul 30;41(30):9269-78. PMID:12135347
- ↑ 4.0 4.1 Grant KL, Klinman JP. Evidence that both protium and deuterium undergo significant tunneling in the reaction catalyzed by bovine serum amine oxidase. Biochemistry. 1989 Aug 8;28(16):6597-605. PMID:2790014
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Raymond Lyle, Alexander Berchansky, OCA, Jaime Prilusky