Delta-endotoxin
From Proteopedia
(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load="2rci" size=" | + | <StructureSection load="2rci" size="350" color="" spin="on" Scene="Cyt2Ba/Cartoon_spectrum/2" caption="''Bacillus thuringiensis'' δ-endotoxin, [[2rci]]" > |
__NOTOC__ | __NOTOC__ | ||
[[Image:Cyt_trim.jpg|left|150px]] | [[Image:Cyt_trim.jpg|left|150px]] | ||
| - | '''Delta-endotoxin''' named also Cyt or Cry, is a pore-forming toxin produced by Bacillus thuringiensis. It is used as an insecticide. Upon ingestion by insect, δ-endotoxin is cleaved, binds to the gut epithelium and forms cation channels. This causes cell lysis and death. For a list of various toxins see [[Toxins]]. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''Delta-endotoxin''' named also Cyt or [[Pesticidal crystal protein|Cry]], is a pore-forming toxin produced by ''Bacillus thuringiensis''. It is used as an insecticide. Upon ingestion by insect, δ-endotoxin is cleaved, binds to the gut epithelium and forms cation channels. This causes cell lysis and death. For a list of various toxins see [[Toxins]]. | ||
| + | See also [[Journal:JMB:1|Cyt1Aa Toxin: High Resolution Structure Reveals Implications for its Membrane-Perforating Function]] | ||
{{Clear}} | {{Clear}} | ||
| Line 12: | Line 34: | ||
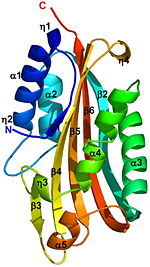
A remarkable similarity is observed between the structures of the endogenously cleaved Cyt2Ba <scene name='Cyt2Ba/Cyt2ba_monomer/2'>monomer</scene> <font color='gray'><b>(gray)</b></font> and the <scene name='Cyt2Ba/Alignment/2'>corresponding region</scene> <font color='red'><b>(red)</b></font> within the inactive protoxin <scene name='Cyt2Ba/Dimer/2'>dimer</scene> of Cyt2Aa ([[1cby]], monomers <font color='red'><b>A</b></font> and <font color='blue'><b>B</b></font> of Cyt2Aa shown <font color='red'><b>red</b></font> and <font color='blue'><b>blue</b></font>, respectively, the N- and C-termini are shown in spacefilling representation). Although, [[1cby]] is a 1 chain structure, the biological relevant molecule for [[1cby]] can be assembled from the contents of the deposited coordinates by the application of crystallographic symmetry operations to give a dimer. It can be [http://www.ebi.ac.uk/pdbe/pqs/macmol/1cby.mmol downloaded]. Each monomer of Cyt2Aa ([[1cby]]), consists of an additional β-strand at its N-terminus and an additional α-helix at its C-terminus compared to the cleaved Cyt2Ba. The <scene name='Cyt2Ba/Dimer_mesh/12'>dimer interface</scene> of Cyt2Aa is held together by the intertwined N-terminal strands from both monomers. The cleavage of Cyt2Aa <scene name='Cyt2Ba/Dimer_mes/1'>removes</scene> the N- and C-terminal segments, prevents dimer formation and releases an <scene name='Cyt2Ba/Monomer_toxin/4'> active toxin monomer</scene>. Similarly, in Cyt2Ba the proteolysis causes the removal of 34 amino acids at its N-terminus and 28 or 30 residues at its C-terminus forming the crystallized toxic monomer. | A remarkable similarity is observed between the structures of the endogenously cleaved Cyt2Ba <scene name='Cyt2Ba/Cyt2ba_monomer/2'>monomer</scene> <font color='gray'><b>(gray)</b></font> and the <scene name='Cyt2Ba/Alignment/2'>corresponding region</scene> <font color='red'><b>(red)</b></font> within the inactive protoxin <scene name='Cyt2Ba/Dimer/2'>dimer</scene> of Cyt2Aa ([[1cby]], monomers <font color='red'><b>A</b></font> and <font color='blue'><b>B</b></font> of Cyt2Aa shown <font color='red'><b>red</b></font> and <font color='blue'><b>blue</b></font>, respectively, the N- and C-termini are shown in spacefilling representation). Although, [[1cby]] is a 1 chain structure, the biological relevant molecule for [[1cby]] can be assembled from the contents of the deposited coordinates by the application of crystallographic symmetry operations to give a dimer. It can be [http://www.ebi.ac.uk/pdbe/pqs/macmol/1cby.mmol downloaded]. Each monomer of Cyt2Aa ([[1cby]]), consists of an additional β-strand at its N-terminus and an additional α-helix at its C-terminus compared to the cleaved Cyt2Ba. The <scene name='Cyt2Ba/Dimer_mesh/12'>dimer interface</scene> of Cyt2Aa is held together by the intertwined N-terminal strands from both monomers. The cleavage of Cyt2Aa <scene name='Cyt2Ba/Dimer_mes/1'>removes</scene> the N- and C-terminal segments, prevents dimer formation and releases an <scene name='Cyt2Ba/Monomer_toxin/4'> active toxin monomer</scene>. Similarly, in Cyt2Ba the proteolysis causes the removal of 34 amino acids at its N-terminus and 28 or 30 residues at its C-terminus forming the crystallized toxic monomer. | ||
The crystal structure of monomeric Cyt2Ba is the first structure of a [http://en.wikipedia.org/wiki/Toxicity toxic] form of the Cyt family. Its structure is [http://en.wikipedia.org/wiki/Homology_(biology) homologous] to the corresponding region of Cyt2Aa and to that of VVA2. This structural comparison shows that the toxicity of Cyt2Ba, Cyt2Aa and VVA2 is an inherent property of the monomer and not the result of secondary structure rearrangement upon cleavage. Solving the 3D structure of these proteins extends the knowledge of the cytolytic machinery of pore-forming toxins and helps in designing novel membrane-active cytotoxins. | The crystal structure of monomeric Cyt2Ba is the first structure of a [http://en.wikipedia.org/wiki/Toxicity toxic] form of the Cyt family. Its structure is [http://en.wikipedia.org/wiki/Homology_(biology) homologous] to the corresponding region of Cyt2Aa and to that of VVA2. This structural comparison shows that the toxicity of Cyt2Ba, Cyt2Aa and VVA2 is an inherent property of the monomer and not the result of secondary structure rearrangement upon cleavage. Solving the 3D structure of these proteins extends the knowledge of the cytolytic machinery of pore-forming toxins and helps in designing novel membrane-active cytotoxins. | ||
| + | |||
| + | The toxicity to insects of the gram-positive bacterium ''Bacillus thuringiensis'', widely used as a biological alternative to chemical pesticides, is due to δ-endotoxic crystals comprised of a series of proteins that react with the cells lining the larval midgut of susceptible insects. The insecticidal proteins are produced during sporulation and classified into two families of membrane perforating toxins, Crystal (Cry) and Cytolytic (Cyt), that are packed into a para-crystalline structure. Following ingestion by an insect of its host range, the Cry and Cyt toxic crystals are solubilized and their pro-toxins are cleaved by the alkaline-active digestive enzymes at the high pH prevailing in the larval midgut. The activated Cry toxins bind to specific protein receptors located on the host cell surface, oligomerize and insert into the membrane, forming lytic pores that cause cell swelling and lysis. In contrast, Cyt toxins do not bind specific receptors but act non-specifically by direct interaction with membrane lipids. However, there is assumption that the toxicity of Cyt1A may be related to the specific unsaturated fatty acid composition of lipids in the midgut epithelial cells of dipteran insects. The two Cyt and Cry families share no common sequence or structural resemblance. They have distinct secondary structures: the α-helical regions of the Cry toxins form the trans-membrane pore, whereas Cyt toxins are presumed to be inserted into the membrane by a β-barrel composed of β-sheet hairpins from each monomer. The activated monomeric form of Cyt1Aa, the most toxic Cyt family member, was isolated and crystallized, and its structure was determined at 2.2 Å resolution (PDB code [[3ron]]). Cyt1Aa adopts a <scene name='Journal:JMB:1/Cv/2'>typical cytolysin fold</scene> containing a <scene name='Journal:JMB:1/Cv/4'>beta-sheet</scene> <span style="color:yellow;background-color:black;font-weight:bold;">(yellow)</span> held by <scene name='Journal:JMB:1/Cv/5'>two surrounding alpha-helical layers</scene> <font color='red'><b>(red)</b></font>. | ||
| + | The conventional model for the Cyt proteins suggests that the monomer undergoes conformational changes, such that <scene name='Journal:JMB:1/Cv/6'>upon membrane contact the two outer alpha-helical layers swing away from the beta-sheet that inserted into the membrane</scene> <font color='blue'><b>(sites of swing labeled in blue)</b></font>. Oligomerization of Cyt monomers on the cell membrane forming β-barrel pores. | ||
| + | Cyt1Aa <span style="color:lime;background-color:black;font-weight:bold;">(lime)</span>, like other Cyt family members, also has a <scene name='Journal:JMB:1/Cv/8'>fold similar to that of the toxic Volvatoxin (VVA2)</scene> (<font color='cyan'><b>cyan</b></font>, PDB code [[1vcy]]) and the non-toxic virulence factor Evf (PDB code [[2w3y]]) despite their very low sequence identity. While, Evf is covalently bound to palmitate, none of the Cyt family members contain a palmitoylated Cys residue. The structural homology between Cyt1Aa and Evf enabled the identification of a <scene name='Journal:JMB:1/Cv/10'>putative fatty acid binding site in Cyt1Aa between the sheet formed by beta4, beta6-beta8 and helices alpha3-alpha5</scene>. The Cyt1Aa structure displays the <scene name='Journal:JMB:1/Cv/13'>conserved hydrophobic residues</scene> <font color='magenta'><b>(colored in magenta)</b></font> pointing towards the putative lipid-binding pocket. We suggest that in Evf, the covalently bound lipid “locks” the helical layer to the β-sheet and prevents the conformational changes necessary for membrane insertion, explaining its observed non-toxicity. On the other hand, the absence of the lipid in Cyt1Aa enables its flexibility and allows the conformational changes of the two surrounding α-helical layers of Cyt1Aa necessary for exposing the hydrophobic β-sheet which is necessary prior to their membrane insertion and perforation. | ||
| + | In attempt to understand why Cyt1Ca is non-toxic, we performed a comparative sequence analysis of all known Cyt1 family members revealing that Cyt1Ca is the most divergent. The residues that are conserved in Cyt1Aa, Cyt1Ab and Cyt1Ba but differ in Cyt1Ca are located on the α-helical layers and on strands β1, β4 and β5 which have been proposed to undergo conformational changes upon membrane binding. The contribution of these residues to the lack of toxicity of Cyt1Ca was supported by the finding that mutating three of these non-conserved residues, Q154, Q164, and G240 in Cyt1Ca to the corresponding charged and exposed residues in Cyt1Aa, K154, E164, and D240 respectively, restored partial antibacterial though not larvicidal activities indicating their importance. We suggest that the lack of its toxicity may also be related to its lack of flexibility. This is supported by the finding that substitution of Q225 in Cyt1Ca to the corresponding conserved K225 in Cyt1Aa, does not restore activity. This residue is located on β8, which is part of the sheet thought to insert into the membrane. We postulate that the location of the non-conserved residues in Cyt1Ca may render this protein unable to undergo the conformational changes associated with membrane insertion, thereby explaining its non-toxicity. | ||
| + | Cyt1Aa synergizes activities of Cry11Aa. Two binding epitopes of Cyt1Aa, <scene name='Journal:JMB:1/Cv1/1'>196-EIKVSAVKE-204</scene> (locating on β7 and α6) and <scene name='Journal:JMB:1/Cv1/2'>220-NIQSLKFAQ-228</scene> (locating on β8), were found to be involved in the binding interaction with Cry11Aa. Both regions are mostly embedded, with only <scene name='Journal:JMB:1/Cv1/3'>200-SAVKE-204</scene> exposed <font color='blueviolet'><b>(colored in blueviolet)</b></font>. The role of these epitopes was confirmed by heterologous competition assays using synthetic peptides. corresponding to these regions and by site directed mutagenesis. In particular, <scene name='Journal:JMB:1/Cv/16'>three single residues, K198, E204 and K225 within these two segments were shown to be involved in the interaction between these two proteins which in turn explain the synergism between them</scene>. Recently it has been shown that mutation of these Cyt1Aa residues affect its binding and synergism with Cry4Ba as well. Interestingly, these three residues are charged in most of the Cyt1 family members, whereas in the Cyt2 family and in Cyt1Ca, which presumably do not bind Cry11Aa, they are polar (T198, Q204 and T225 respectively in Cyt2Ba). Thus, it seems reasonable that synergism and binding of Cyt1Aa to Cry11Aa or to Cry4Ba depend on specific interactions between these toxins, which involve these residues. We suggest that the reduced charge on the Cyt2 protein members and on Cyt1Ca may be sufficient to abrogate binding to Cry11Aa. It was suggested that mutating these residues in other Cyt proteins to the corresponding Cyt1Aa charged residues might introduce binding sites and induce synergism with Cry toxins. This strategy could be used as a tool to overcome Cry-resistance in the midgut membrane of resistant insects. A sequential mechanism has been proposed by which Cyt1Aa initially undergoes conformational changes to insert its β-sheet into the membrane following binding of Cry11Aa via the two Cyt1Aa binding epitopes resulting in insertion of Cry11Aa into the mosquito membranes. Mapping the three charged residues on the Cyt1Aa structure revealed that while all three residues are exposed to the surface of the protein, they all reside on regions of the toxin which presumably are inserted into the membrane (K198 and E204 are located on β7 and α6, and K225 is part of β8). We therefore, can't out rule an alternative mechanism by which Cyt1Aa binds Cry11Aa using these exposed charged residues prior to its membrane insertion. Thus, the action of Cyt1Aa alone or as a receptor for Cry11Aa may involve different mechanisms. | ||
| + | The pattern of the hemolytic activity of Cyt1Aa presented here (resembling that of pore-forming agents), while differing from that imposed by ionic and nonionic detergents, further supports the pore-forming model by which conformational changes occur prior to membrane insertion and perforation. | ||
</StructureSection> | </StructureSection> | ||
| Line 20: | Line 49: | ||
[[3eb7]] – BtCry8Ea1 – ''Bacillus thuringiensis''<br /> | [[3eb7]] – BtCry8Ea1 – ''Bacillus thuringiensis''<br /> | ||
| - | [[2rci]] – BtCytBa<br /> | + | [[2rci]], [[3ron]] – BtCytBa<br /> |
| - | [[4arx]] – BtCry1Ac<br /> | + | [[1ciy]], [[8w7n]] – BtCry1Aa<br /> |
| + | [[7qx4]], [[7qx5]], [[7qx6]] – BtCry1Aa (mutant)<br /> | ||
| + | [[4arx]], [[6t14]], [[6t19]], [[6t1a]] – BtCry1Ac<br /> | ||
| + | [[4w8j]], [[6t1c]] – BtCry1Ac (mutant)<br /> | ||
[[4ary]] – BtCry1Ac + GalNac<br /> | [[4ary]] – BtCry1Ac + GalNac<br /> | ||
| + | [[6dj4]] – BtCry1A.105 <br /> | ||
| + | [[6wpc]] – BtCry1A.2 <br /> | ||
| + | [[6owk]] – BtCry1Be <br /> | ||
| + | [[6ovb]] – BtCry1Da <br /> | ||
| + | [[9buv]] – BtCry1Fa (mutant)<br /> | ||
| + | [[1i5p]] – BtCry2Aa<br /> | ||
[[4qx0]], [[4qx1]], [[4qx2]], [[4qx3]] – BtCry3Aa<br /> | [[4qx0]], [[4qx1]], [[4qx2]], [[4qx3]] – BtCry3Aa<br /> | ||
[[2c9k]] – BtCry4A residues 68-679<br /> | [[2c9k]] – BtCry4A residues 68-679<br /> | ||
| Line 28: | Line 66: | ||
[[4d8m]] - BtCry4Ba residues 112-698<br /> | [[4d8m]] - BtCry4Ba residues 112-698<br /> | ||
[[4moa]] - BtCry4Ba residues 40-641 (mutant)<br /> | [[4moa]] - BtCry4Ba residues 40-641 (mutant)<br /> | ||
| + | [[5ghe]], [[5kuc]], [[5kud]] – BtCry6Aa<br /> | ||
[[1dlc]] - BtCryIiia residues 61-644<br /> | [[1dlc]] - BtCryIiia residues 61-644<br /> | ||
[[1ji6]] - BtCryIiib residues 64-652<br /> | [[1ji6]] - BtCryIiib residues 64-652<br /> | ||
[[1cby]] - BtCytB residues 1-259<br /> | [[1cby]] - BtCytB residues 1-259<br /> | ||
| - | [[3ron]] – BtCry1aa<br /> | ||
[[2mlw]] – CytC 1BA – ''Dickeya dadantii''<br /> | [[2mlw]] – CytC 1BA – ''Dickeya dadantii''<br /> | ||
Current revision
| |||||||||||
3D Structures of δ-endotoxin
Updated on 01-April-2025
3eb7 – BtCry8Ea1 – Bacillus thuringiensis
2rci, 3ron – BtCytBa
1ciy, 8w7n – BtCry1Aa
7qx4, 7qx5, 7qx6 – BtCry1Aa (mutant)
4arx, 6t14, 6t19, 6t1a – BtCry1Ac
4w8j, 6t1c – BtCry1Ac (mutant)
4ary – BtCry1Ac + GalNac
6dj4 – BtCry1A.105
6wpc – BtCry1A.2
6owk – BtCry1Be
6ovb – BtCry1Da
9buv – BtCry1Fa (mutant)
1i5p – BtCry2Aa
4qx0, 4qx1, 4qx2, 4qx3 – BtCry3Aa
2c9k – BtCry4A residues 68-679
1w99 - BtCry4Ba residues 84-641
4d8m - BtCry4Ba residues 112-698
4moa - BtCry4Ba residues 40-641 (mutant)
5ghe, 5kuc, 5kud – BtCry6Aa
1dlc - BtCryIiia residues 61-644
1ji6 - BtCryIiib residues 64-652
1cby - BtCytB residues 1-259
2mlw – CytC 1BA – Dickeya dadantii
References
- Li J, Koni PA, Ellar DJ. Structure of the mosquitocidal delta-endotoxin CytB from Bacillus thuringiensis sp. kyushuensis and implications for membrane pore formation. J Mol Biol. 1996 Mar 22;257(1):129-52. PMID:8632451 doi:http://dx.doi.org/10.1006/jmbi.1996.0152
- Cohen S, Dym O, Albeck S, Ben-Dov E, Cahan R, Firer M, Zaritsky A. High-resolution crystal structure of activated Cyt2Ba monomer from Bacillus thuringiensis subsp. israelensis. J Mol Biol. 2008 Jul 25;380(5):820-7. Epub 2008 May 11. PMID:18571667 doi:10.1016/j.jmb.2008.05.010