Diguanylate cyclase
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
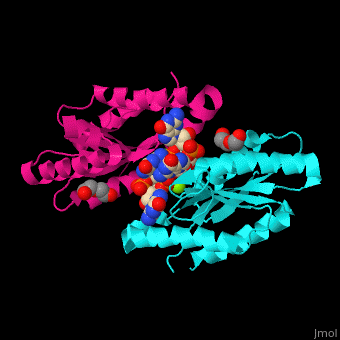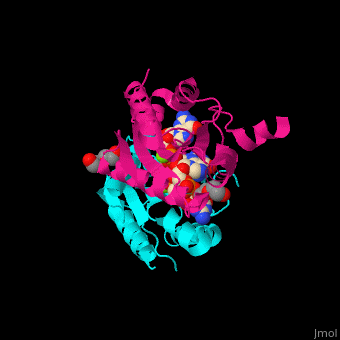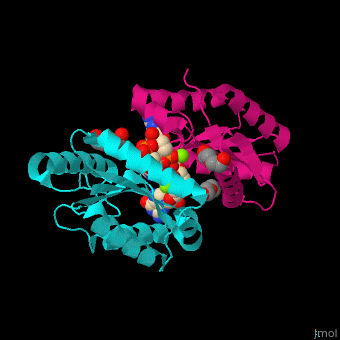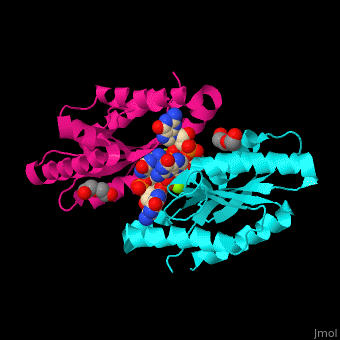
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='Diguanylate cyclase GGDEF domain complex with cyclic di-GMP, Mg+2 ion (green) and PEG (PDB code [[3qyy]]).' scene='67/677040/Cv/1'> |
| Line 5: | Line 5: | ||
'''Diguanylate cyclase''' (DGC) catalyzes the conversion of GTP to [[C-di-GMP]] and diphosphate. <ref>PMID:17697992</ref> For more details see<br /> | '''Diguanylate cyclase''' (DGC) catalyzes the conversion of GTP to [[C-di-GMP]] and diphosphate. <ref>PMID:17697992</ref> For more details see<br /> | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[C-di-GMP signaling]]. | [[C-di-GMP signaling]]. | ||
| Line 16: | Line 14: | ||
== Structural highlights == | == Structural highlights == | ||
| - | DGC contains the sequence <scene name='67/677040/Cv/ | + | DGC contains the sequence <scene name='67/677040/Cv/4'>motif GGDEF</scene> in its active site. <scene name='67/677040/Cv/5'>Cyclic di-GMP and Mg+2 ions interact with two monomers</scene> of DGC.<ref>PMID:22120736</ref> |
</StructureSection> | </StructureSection> | ||
| Line 26: | Line 24: | ||
*DGC | *DGC | ||
| - | **[[4urg]] – TmDGC GGDEF domain – ''Thermotoga maritima''<br /> | + | **[[4urg]], [[6khu]] – TmDGC GGDEF domain 82-248 – ''Thermotoga maritima''<br /> |
**[[4urq]] – TmDGC GGDEF domain (mutant)<br /> | **[[4urq]] – TmDGC GGDEF domain (mutant)<br /> | ||
**[[3ign]] – MaDGC GGDEF domain – ''Marinobacter aquaeolei''<br /> | **[[3ign]] – MaDGC GGDEF domain – ''Marinobacter aquaeolei''<br /> | ||
**[[3h9w]] – MaDGC N terminal<br /> | **[[3h9w]] – MaDGC N terminal<br /> | ||
| - | **[[ | + | **[[8wcn]] – PaDGC + GMPCPP – ''Pseudomonas aeruginosa'' – Cryo EM<br /> |
| + | **[[4zmu]] – PaDGC 1-341 <br /> | ||
| + | **[[3hvw]], [[4iob]] – DGC GGDEF domain 254-414<br /> | ||
| + | **[[7a7e]] – PaDGC GGDEF domain (mutant)<br /> | ||
| + | **[[4zmm]] – PaDGC GGDEF domain + C-diGMP<br /> | ||
| + | **[[5m3c]] – PaDGC GGDEF domain + GTP<br /> | ||
| + | **[[7e6g]] – PaDGC GGDEF domain + SiaC<br /> | ||
**[[3n53]] – DGC – ''Pelobacter carbinolicus''<br /> | **[[3n53]] – DGC – ''Pelobacter carbinolicus''<br /> | ||
| - | **[[ | + | **[[5h5o]] – XcDGC residues 1-150 – ''Xanthomonas campestris''<br /> |
**[[3qyy]] – XcDGC GGDEF domain + cyclic di-GMP <br /> | **[[3qyy]] – XcDGC GGDEF domain + cyclic di-GMP <br /> | ||
**[[4urs]] – TmDGC GGDEF domain + cyclic di-GMP<br /> | **[[4urs]] – TmDGC GGDEF domain + cyclic di-GMP<br /> | ||
**[[3t9o]] – EcDGC YDEH CZB domain – ''Escherichia coli''<br /> | **[[3t9o]] – EcDGC YDEH CZB domain – ''Escherichia coli''<br /> | ||
| - | **[[3tvk]] – EcDGC YDEH GGDEF domain <br /> | + | **[[3tvk]], [[4zve]], [[4zvg]], [[4zvh]] – EcDGC YDEH GGDEF domain <br /> |
**[[4h54]] – EcDGC YDEH (mutant) <br /> | **[[4h54]] – EcDGC YDEH (mutant) <br /> | ||
| + | **[[4zva]], [[4zvb]] – EcDGC globin domain <br /> | ||
| + | **[[4zvc]], [[4zvd]] – EcDGC mid domain <br /> | ||
| + | **[[4zvf]] – EcDGC GGDEF domain 297-460 + GTP derivative<br /> | ||
| + | **[[6eib]] – DGC GGDEF domain – ''Vibrio cholerae''<br /> | ||
| + | **[[6et7]] – IdDGC (mutant) - ''Idiomarina''<br /> | ||
| + | **[[5llw]], [[5lly]] – IdDGC + phytochromobilin <br /> | ||
| + | **[[5llx]] – IdDGC + phytochromobilin + GTP <br /> | ||
| + | **[[6saw]], [[6sax]] – IdDGC chromophore-binding domain 3-312<br /> | ||
| + | **[[6hbz]] – DGCB – ''Bdellovibrio bacteriovorus''<br /> | ||
| + | **[[6tts]] – CvDGCB GGDEF domain 176-353 + C-diGMP – ''Caulobacter vibrioides'' <br /> | ||
| + | **[[6ttr]] – CvDGCB GGDEF+coiled-coil domains 153-353 (mutant) + C-diGMP <br /> | ||
| + | **[[7k5n]] – DGC LBD domain 39-286 + Pro – ''Aeromonas caviae''<br /> | ||
| + | **[[8px4]] – DGC – ''Leptospira interogans''<br /> | ||
| + | **[[8c05]] – DGC + FMN – ''Methylotenera''<br /> | ||
| - | *Response regulator PleD | ||
| - | |||
| - | **[[1w25]], [[2wb4]] – CvDGC + cyclic di-GMP – ''Caulobacter vibrioides''<br /> | ||
| - | **[[2v0n]] – CvDGC + cyclic di-GMP + GTP<br /> | ||
}} | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D Structures of diguanylate cyclase
Updated on 02-April-2025
References
- ↑ Stock AM. Diguanylate cyclase activation: it takes two. Structure. 2007 Aug;15(8):887-8. PMID:17697992 doi:http://dx.doi.org/10.1016/j.str.2007.07.003
- ↑ Yang CY, Chin KH, Chuah ML, Liang ZX, Wang AH, Chou SH. The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)(2) at the active site. Acta Crystallogr D Biol Crystallogr. 2011 Dec;67(Pt 12):997-1008. Epub 2011 Nov, 18. PMID:22120736 doi:10.1107/S090744491104039X