We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Ectonucleoside triphosphate diphosphohydrolase
From Proteopedia
(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
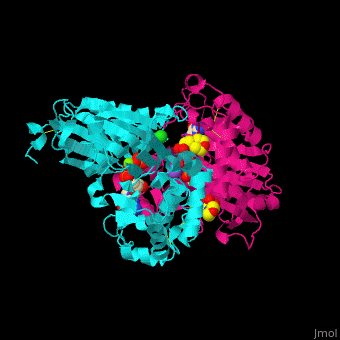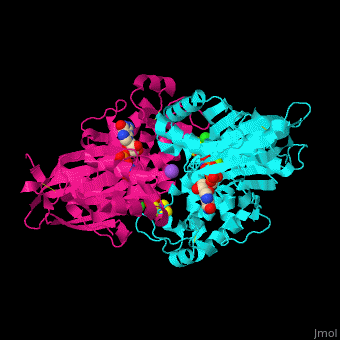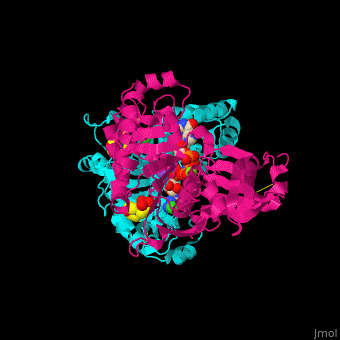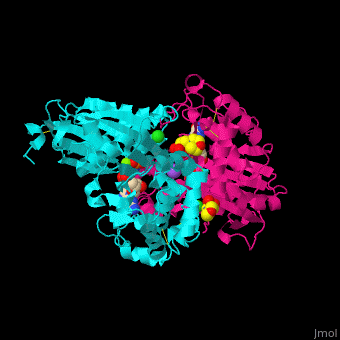
| - | <StructureSection load='4brg' size=' | + | <StructureSection load='4brg' size='400' side='right' caption='Structure of NTDPase1 complex with GMPPNP, Na+ (purple), Cl- (large green), Mg+2 (small green) ions (PDB code [[4brg]]).' scene='59/597004/Cv/1'> |
| - | == Function == | ||
| - | '''Ectonucleoside triphosphate diphosphohydrolase (NTPDase)''' hydrolyzes nucleoside 5’-triphosphate to nucleoside 5’-phosphate and 2 phosphates. NTPDase can hydrolyze ATP and ADP. NTPDase has an important role in regulating neutrophil chemotaxis. NTPDase 1,2,3,8 are cell surface enzymes with catalytic sites facing extracellularly while other NTPDases act intracellularly. NTPDase require Ca+2 or Mg+2 for their activity. The NTPDases can be differentiated according to their substrate specificity, divalent cation usage and product formation. | + | '''Ectonucleoside triphosphate diphosphohydrolase (NTPDase)''' hydrolyzes nucleoside 5’-triphosphate to nucleoside 5’-phosphate and 2 phosphates. NTPDase can hydrolyze ATP and ADP. NTPDase has an important role in regulating neutrophil chemotaxis. NTPDase 1,2,3,8 are cell surface enzymes with catalytic sites facing extracellularly while other NTPDases act intracellularly. NTPDase require Ca<sup>+2</sup> or Mg<sup>+2</sup> for their activity. The NTPDases can be differentiated according to their substrate specificity, divalent cation usage and product formation.<ref>PMID:23830739</ref> |
| + | *'''NTPDase 1''' hydrolyzes ATP released by neutrophils<ref>PMID:18713747</ref>. | ||
| + | *'''NTPDase 2''' hydrolyzes extracellular ATP <ref>PMID:35593054</ref>. | ||
| + | *'''NTPDase 3''' hydrolyzes ATP in pancreatic beta-cells<ref>PMID:24085034</ref>. | ||
| + | *'''NTPDase 4''' hydrolyzes ATP in Golgi bodies<ref>PMID:32767432</ref>. | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==3D structures of ectonucleoside triphosphate diphosphohydrolase == | ==3D structures of ectonucleoside triphosphate diphosphohydrolase == | ||
| Line 18: | Line 17: | ||
*NTPDase1 | *NTPDase1 | ||
| + | **[[3zx3]] – rNTPDase extracellular domain - rat<br /> | ||
| + | **[[3zx0]], [[3zx2]] – rNTPDase extracellular domain + polyoxometallates <br /> | ||
| + | **[[5u7p]] – cNTPDase - clover<br /> | ||
| + | **[[5u7v]] – cNTPDase + AMP <br /> | ||
| + | **[[5u7w]] – cNTPDase + adenine <br /> | ||
| + | **[[5u7x]] – NTPDase - ''Vigna unguiculata''<br /> | ||
**[[3aap]] – LpNTPDase – ''Legionella pneumophila''<br /> | **[[3aap]] – LpNTPDase – ''Legionella pneumophila''<br /> | ||
**[[3aaq]] – LpNTPDase + inhibitor<br /> | **[[3aaq]] – LpNTPDase + inhibitor<br /> | ||
**[[3aar]] – LpNTPDase + AMPPNP<br /> | **[[3aar]] – LpNTPDase + AMPPNP<br /> | ||
| - | **[[3zx3]] – rNTPDase extracellular domain - rat<br /> | ||
| - | **[[3zx0]], [[3zx2]] – rNTPDase extracellular domain + polyoxometallates <br /> | ||
**[[4jep]] – TgNTPDase – ''Toxoplasma gondii''<br /> | **[[4jep]] – TgNTPDase – ''Toxoplasma gondii''<br /> | ||
**[[4a5b]] – TgNTPDase (mutant) <br /> | **[[4a5b]] – TgNTPDase (mutant) <br /> | ||
| Line 34: | Line 37: | ||
*NTPDase1 closed form | *NTPDase1 closed form | ||
| - | **[[4br9]] – LpNTPDase <br /> | + | **[[4br9]], [[4brp]] – LpNTPDase <br /> |
**[[4bra]] – LpNTPDase + AMPPNP<br /> | **[[4bra]] – LpNTPDase + AMPPNP<br /> | ||
**[[4brc]] – LpNTPDase + adenosine-imido-diphosphate<br /> | **[[4brc]] – LpNTPDase + adenosine-imido-diphosphate<br /> | ||
| Line 57: | Line 60: | ||
**[[3cj7]] – rNTPDase extracellular domain + AMP<br /> | **[[3cj7]] – rNTPDase extracellular domain + AMP<br /> | ||
**[[3cj9]] – rNTPDase extracellular domain + AMP + phosphate<br /> | **[[3cj9]] – rNTPDase extracellular domain + AMP + phosphate<br /> | ||
| - | **[[3cja]] – rNTPDase extracellular domain + AMPPNP<br /> | + | **[[3cja]], [[4br5]] – rNTPDase extracellular domain + AMPPNP<br /> |
**[[4bqz]] – rNTPDase extracellular domain + GMPPNP<br /> | **[[4bqz]] – rNTPDase extracellular domain + GMPPNP<br /> | ||
**[[4br2]] – rNTPDase extracellular domain + UMPPNP<br /> | **[[4br2]] – rNTPDase extracellular domain + UMPPNP<br /> | ||
| Line 70: | Line 73: | ||
**[[4a5a]], [[4kh4]] – TgNTPDase (mutant) + AMPPNP<br /> | **[[4a5a]], [[4kh4]] – TgNTPDase (mutant) + AMPPNP<br /> | ||
**[[4kh5]] – TgNTPDase (mutant) + adenosine-imido-diphosphate<br /> | **[[4kh5]] – TgNTPDase (mutant) + adenosine-imido-diphosphate<br /> | ||
| + | |||
| + | *NTPDase4 | ||
| + | |||
| + | **[[6wg5]] – hNTPDase <br /> | ||
}} | }} | ||
Current revision
| |||||||||||
3D structures of ectonucleoside triphosphate diphosphohydrolase
Updated on 06-May-2025
References
- ↑ Zebisch M, Krauss M, Schafer P, Lauble P, Strater N. Crystallographic Snapshots along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases. Structure. 2013 Jul 2. pii: S0969-2126(13)00200-1. doi:, 10.1016/j.str.2013.05.016. PMID:23830739 doi:10.1016/j.str.2013.05.016
- ↑ Corriden R, Chen Y, Inoue Y, Beldi G, Robson SC, Insel PA, Junger WG. Ecto-nucleoside triphosphate diphosphohydrolase 1 (E-NTPDase1/CD39) regulates neutrophil chemotaxis by hydrolyzing released ATP to adenosine. J Biol Chem. 2008 Oct 17;283(42):28480-6. PMID:18713747 doi:10.1074/jbc.M800039200
- ↑ Dragic M, Mihajlovic K, Adzic M, Jakovljevic M, Kontic MZ, Mitrović N, Laketa D, Lavrnja I, Kipp M, Grković I, Nedeljkovic N. Expression of Ectonucleoside Triphosphate Diphosphohydrolase 2 (NTPDase2) Is Negatively Regulated Under Neuroinflammatory Conditions In Vivo and In Vitro. ASN Neuro. 2022 Jan-Dec;14:17590914221102068. PMID:35593054 doi:10.1177/17590914221102068
- ↑ Syed SK, Kauffman AL, Beavers LS, Alston JT, Farb TB, Ficorilli J, Marcelo MC, Brenner MB, Bokvist K, Barrett DG, Efanov AM. Ectonucleotidase NTPDase3 is abundant in pancreatic β-cells and regulates glucose-induced insulin secretion. Am J Physiol Endocrinol Metab. 2013 Nov 15;305(10):E1319-26. PMID:24085034 doi:10.1152/ajpendo.00328.2013
- ↑ Gorelik A, Labriola JM, Illes K, Nagar B. Crystal structure of the nucleotide-metabolizing enzyme NTPDase4. Protein Sci. 2020 Aug 6. doi: 10.1002/pro.3926. PMID:32767432 doi:http://dx.doi.org/10.1002/pro.3926