Journal:Acta Cryst F:S2053230X25007034
From Proteopedia
(Difference between revisions)

| (57 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene='10/1087242/ | + | <StructureSection load='' size='450' side='right' scene='10/1087242/019_fig_2a_png/1' caption='The structure of HNL6V ([[8euo]]), an α/β-hydrolase fold enzyme. The catalytic triad (S80, H235, D207) orange sticks with the Oɣ of Ser80 in red at the center of the figure. The pink spheres show the Cɑ’s of the seven substitutions (T11G, E79H, C81L, H103V, D104A, G176S, K236M) the were engineered to convert hydroxynitrile lyase from rubber trees (HbHNL) to make it more structurally similar to SABP2, an esterase from tobacco that shares 44% sequence identity.'> |
===Crystal structure of a seven-substitution mutant of hydroxynitrile lyase from rubber tree=== | ===Crystal structure of a seven-substitution mutant of hydroxynitrile lyase from rubber tree=== | ||
| - | <big> | + | <big>Colin T. Pierce, Lauren R. Greenberg, Meghan E. Walsh, Ke Shi, Drenen J. Mageea, Hideki |
| + | Aihara, Wendy Gordon, Robert L. Evans III and Romas J. Kazlauskas</big> <ref>doi: 10.1107/S2053230X25007034</ref> | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
The α/β-hydrolase fold superfamily serves as an excellent example of functional divergence—enzymes sharing nearly identical structures yet catalyzing different reactions. While most members of this family are esterases that hydrolyze ester bonds, hydroxynitrile lyases (HNLs) in this family have evolved to catalyze the reversible cleavage of cyanohydrins into aldehydes and hydrogen cyanide. Despite this functional divergence, both enzyme types retain the canonical Ser-His-Asp catalytic triad, raising questions about how active site geometry determines the catalytic mechanism. | The α/β-hydrolase fold superfamily serves as an excellent example of functional divergence—enzymes sharing nearly identical structures yet catalyzing different reactions. While most members of this family are esterases that hydrolyze ester bonds, hydroxynitrile lyases (HNLs) in this family have evolved to catalyze the reversible cleavage of cyanohydrins into aldehydes and hydrogen cyanide. Despite this functional divergence, both enzyme types retain the canonical Ser-His-Asp catalytic triad, raising questions about how active site geometry determines the catalytic mechanism. | ||
| - | The team focused on hydroxynitrile lyase from rubber trees (HbHNL) and attempted to make it more structurally similar to SABP2, an esterase from tobacco that shares 44% sequence identity. By introducing seven strategic mutations that match the corresponding positions in the target esterase, the researchers created <scene name='10/1087242/ | + | The team focused on hydroxynitrile lyase from rubber trees (HbHNL) and attempted to make it more structurally similar to SABP2, an esterase from tobacco that shares 44% sequence identity. By introducing seven strategic mutations that match the corresponding positions in the target esterase, the researchers created <scene name='10/1087242/019_Fig_2a_sticks_pse/1'>HNL6V, a hybrid enzyme</scene> that sits structurally between its parent and target enzymes. Six of the seven substitutions target the catalytic domain within or immediately adjacent to the active site, while the seventh targets the lid domain, also adjacent to the active site. |
| - | The 1.99-Å crystal structure of HNL6V reveals the molecular basis for this engineered transformation. The seven substitutions induced <scene name='10/1087242/ | + | The 1.99-Å crystal structure of HNL6V reveals the molecular basis for this engineered transformation. The seven substitutions induced <scene name='10/1087242/019_fig_02a_no_dist_png/3'>subtle but systematic shifts throughout the protein</scene>, moving the catalytic atoms 0.2-0.8 Å closer to their positions in the target esterase while increasing flexibility in the lid domain—demonstrating that the mutations both repositioned key atoms and fine-tuned protein dynamics. This work demonstrates that evolutionary trajectories between related enzymes can be partially retraced through rational design, offering new strategies for engineering enzymes with intermediate or altered catalytic properties. |
| - | [[Image:019 Fig 3a.triad label.jpg|thumb|left| | + | {| class="wikitable" style="margin:auto" |
| - | + | |- | |
| - | + | |- | |
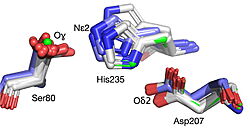
| - | + | | [[Image:019 Fig 3a.triad label.jpg|thumb|left|250px|Best fit overlay of the Cɑ positions of SABP2 structures (three structures, light blue carbons) and HbHNL structures (eighteen structures, white carbons) onto the structure of | |
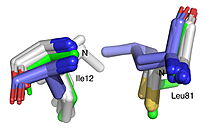
| + | HNL6V (green sticks). The catalytic atoms of the catalytic triad of this α/β-hydrolase fold, in HNL6V (Oɣ of S80, Nε2 of H235, Oδ2 of D207) overlay more closely with the corresponding atoms in the HbHNL structures (S80, H235, A207) than with the corresponding atoms in the SABP2 structures (S81, H238, D210).]] [[Image:019_Fig_3b.oxyanion_label.jpg|thumb|right|210px|Best fit overlay of the Cɑ positions of SABP2 structures (three structures, light blue carbons) and HbHNL structures (eighteen structures, white carbons) onto the structure of HNL6V (green sticks). The oxyanion hole amide nitrogen atoms of I12 and L81 in HNL6V overlay more closely with the corresponding atoms in HbHNL (I12, C81) than with the corresponding atoms in SABP2 (A13 and L82).]] | ||
| + | |} | ||
<b>References</b><br> | <b>References</b><br> | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.