Histone deacetylase
From Proteopedia
(Difference between revisions)
| (37 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
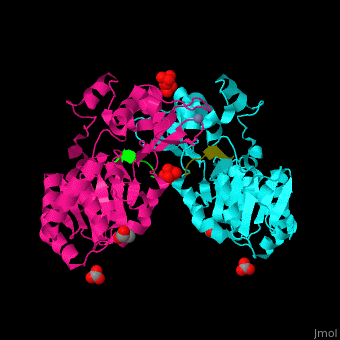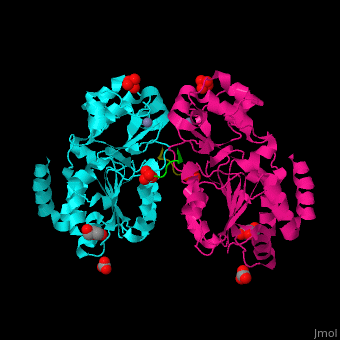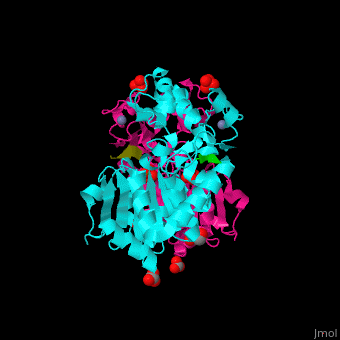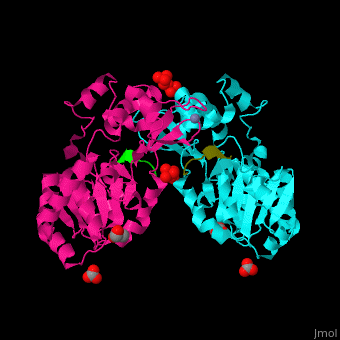
| - | + | <StructureSection load='' size='350' side='right' caption='Human sirtuin-3 complex with peptide containing acetylated lysine, sulfate, carbonate, glycerol and Zn+2 ion (PDB entry [[3glr]])' scene='48/483191/Cv/1'> | |
| + | == Function == | ||
| + | '''Histone deacetylase''' (HDAC) catalyzes the removal of acetyl group from ε-N-acetyl lysine in histones<ref>PMID:23752268</ref>. HDAC contains Zn. DNA expression is regulated by acetylation and de-acetylation. SAHA is a common inhibitor of HDAC. HDAC are classified according to their domain organization into 4 classes. <br /> | ||
| + | * '''HDAC class I''' are homologous to yeast Rpd3.<br /> | ||
| + | * '''HDAC class II''' are homologous to yeast HdaI.<br /> | ||
| + | * '''HDAC class IIa''' lack enzymatic activity<ref>PMID:25244360</ref>. <br /> | ||
| + | * '''HDAC class III''' called '''sirtuin''' - (Silent Information Regulator) (SIRT) are NAD-dependent HDAC. This protein is bifunctional and can act as a deacylase or as mono-ADP-acetyltransferase. <ref>PMID:18249170</ref>.<br /> | ||
| + | * '''HDAC Rpd3''' belongs to class I and is involved in transcriptional repression <ref>PMID:9512514</ref>.<br /> | ||
| + | * '''HDAC Clr (Cryptic Loci Regulator)''' is responsible of deacetylation of Lys residues at the N-terminal of histones .<br /> | ||
| - | + | For HDAC8 see - [[Histone deacetylase 8 (HDAC8)]].<br /> | |
| + | For additional details see<br /> | ||
| + | * [[Understanding of the Recruitment of HDACs by MEF2, Based on Their Structure]]<br /> | ||
| + | * [[Transcription and RNA Processing]]. | ||
| - | + | == Relevance == | |
| + | HDAC inhibitors are used in cancer therapy<ref>PMID:11902574</ref>. Sirtuins play a role in cancer, metabolic activity and neurodegenerative diseases<ref>PMID:17456799</ref>. HDAC inhibitor SAHA or [[Zolinza (Vorinostat)]] is used in the treatment of cutaneous T cell lymphoma<ref>PMID:17962618</ref>. | ||
| + | |||
| + | == Structural highlights == | ||
| + | The biological assembly of Human sirtuin-3 is <scene name='48/483191/Cv/5'>homodimer</scene>. The <scene name='48/483191/Cv/6'>acetylated lysine of the peptide is inserted in an extended conformation</scene> between the 2 domains of SIRT3 - a large Rossman fold, NAD-binding domain and a small Zn-binding one)<ref>PMID:19535340</ref>. Water molecule shown as red sphere. | ||
== 3D Structures of histone deacetylase == | == 3D Structures of histone deacetylase == | ||
| + | [[Histone deacetylase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. PMID:23752268 doi:http://dx.doi.org/10.1038/msb.2013.26
- ↑ Parra M. Class IIa HDACs FEBS J. 2015 May;282(9):1736-44. PMID:25244360 doi:10.1111/febs.13061
- ↑ Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008 Feb;7(2):104-12. doi: 10.1016/j.cmet.2007.11.006. PMID:18249170 doi:http://dx.doi.org/10.1016/j.cmet.2007.11.006
- ↑ Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998 Mar 15;12(6):797-805. PMID:9512514 doi:10.1101/gad.12.6.797
- ↑ Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001 Dec;1(3):194-202. PMID:11902574 doi:http://dx.doi.org/10.1038/35106079
- ↑ Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007 Aug;21(8):1745-55. Epub 2007 Apr 24. PMID:17456799 doi:http://dx.doi.org/10.1210/me.2007-0079
- ↑ Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007 Oct;12(10):1247-52. PMID:17962618 doi:http://dx.doi.org/10.1634/theoncologist.12-10-1247
- ↑ Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, Milne JC, Westphal CH, Perni RB. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem. 2009 Sep 4;284(36):24394-405. Epub 2009 Jun 16. PMID:19535340 doi:10.1074/jbc.M109.014928