Monkeypox DNA Polymerase
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
'''DNA path:''' duplex lies in groove between palm and thumb. Single-stranded 5′ template exits through a channel formed by F8 NTD + Exo and E4, perpendicular to the duplex. | '''DNA path:''' duplex lies in groove between palm and thumb. Single-stranded 5′ template exits through a channel formed by F8 NTD + Exo and E4, perpendicular to the duplex. | ||
| - | '''Residues coordinating catalysis / substrate:''' conserved Asp residues D549 (motif A- F8) and D753 (motif C- A22) coordinate the catalytic metal; Y554 stacks the incoming ribose (steric gate against rNTPs); R634 and K661 stabilize triphosphate. These motifs closely mirror canonical B-family polymerases. | + | '''Residues coordinating catalysis / substrate:''' conserved Asp residues D549 (motif A- F8) and D753 (motif C- A22) coordinate the catalytic metal; Y554 of F8 stacks the incoming ribose (steric gate against rNTPs); R634 and K661 stabilize triphosphate. These motifs closely mirror canonical B-family polymerases. |
---- | ---- | ||
| Line 47: | Line 47: | ||
'''Supporting biochemistry:''' Primer-extension assays show F8 alone is distributive (incorporates <14 nt), while addition/assembly with A22–E4 yields full-length extension (60 nt template) in a concentration-dependent manner; alanine scanning of E4 residues (e.g., W36, R39, N165) confirms residues critical for processivity. | '''Supporting biochemistry:''' Primer-extension assays show F8 alone is distributive (incorporates <14 nt), while addition/assembly with A22–E4 yields full-length extension (60 nt template) in a concentration-dependent manner; alanine scanning of E4 residues (e.g., W36, R39, N165) confirms residues critical for processivity. | ||
---- | ---- | ||
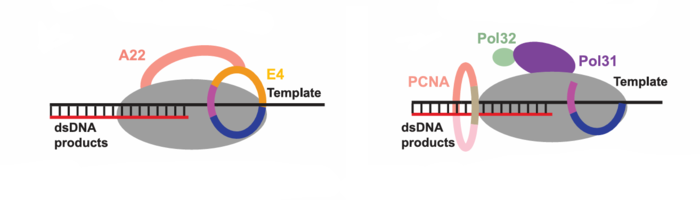
| - | + | The following figure shows two binding modes of processivity factors with polymerases. The processivity factors bound with template in poxvirus function as a “forward sliding clamp” (i) or dsDNA products in eukaryotes as a “backward sliding clamp” (ii). | |
| + | |||
[[Image:MPXV.png|700px]] | [[Image:MPXV.png|700px]] | ||
Revision as of 01:20, 30 November 2025
Your Heading Here (maybe something like 'Structure')
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Yana Fedotova, Eric Martz, Silky Srivastava, Alicia Daeden, Jaime Prilusky, Susana Retamal, Eran Hodis, Wayne Decatur