Monkeypox DNA Polymerase
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
==Overall Architecture== | ==Overall Architecture== | ||
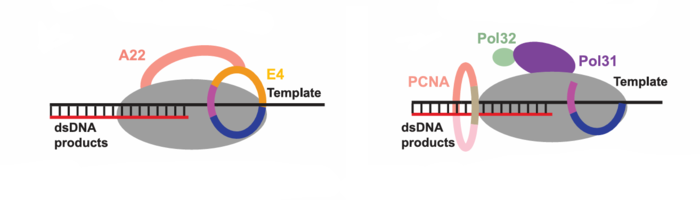
| - | ''' | + | '''Components of holoenzyme:''' 1 <scene name='33/330227/Dna_polymerase_f8/1'>F8</scene> , 1 <scene name='33/330227/A22/1'>A22</scene> , 1 <scene name='33/330227/E4/2'>E4</scene> , <scene name='33/330227/Primer/2'>primer</scene>–<scene name='33/330227/Template/1'>template</scene> DNA , incoming <scene name='33/330227/Dttp/1'>dTTP</scene>. |
'''F8 (polymerase):''' 1004 residues traced (last two residues missing); canonical B-family domains — NTD, 3′–5′ Exonuclease (Exo), palm, fingers, thumb — plus five poxvirus-specific insertions (largest named insert2). | '''F8 (polymerase):''' 1004 residues traced (last two residues missing); canonical B-family domains — NTD, 3′–5′ Exonuclease (Exo), palm, fingers, thumb — plus five poxvirus-specific insertions (largest named insert2). | ||
| - | '''A22 (processivity factor):''' three domains — NTD, Middle (Mid), CTD. The Mid shows structural similarity to ligase adenylylation and OB-fold | + | '''A22 (processivity factor):''' three domains — NTD, Middle (Mid), CTD. The Mid shows structural similarity to ligase adenylylation and OB-fold domains but lacks canonical ligase activity (putative active site is nonfunctional). |
'''E4 (uracil-DNA glycosylase homolog):''' ~218 residues, resembles VACV D4 and contacts F8 Exo directly. | '''E4 (uracil-DNA glycosylase homolog):''' ~218 residues, resembles VACV D4 and contacts F8 Exo directly. | ||
| Line 34: | Line 34: | ||
'''DNA path:''' duplex lies in groove between palm and thumb. Single-stranded 5′ template exits through a channel formed by F8 NTD + Exo and E4, perpendicular to the duplex. | '''DNA path:''' duplex lies in groove between palm and thumb. Single-stranded 5′ template exits through a channel formed by F8 NTD + Exo and E4, perpendicular to the duplex. | ||
| - | '''Residues | + | '''Residues involved:''' conserved Asp residues D549 (motif A- F8) and D753 (motif C- A22) coordinate the catalytic metal; Y554 of F8 stacks the incoming ribose (steric gate against rNTPs); R634 and K661 stabilize triphosphate. These motifs closely mirror canonical B-family polymerases. |
---- | ---- | ||
Revision as of 02:01, 30 November 2025
Structure of DNA Polymerase Holoenzyme of Monkeypox Virus
| |||||||||||
Note
This page was prepared as part of the internal assessment for course BI3323-Aug2025.
Proteopedia Page Contributors and Editors (what is this?)
Yana Fedotova, Eric Martz, Silky Srivastava, Alicia Daeden, Jaime Prilusky, Susana Retamal, Eran Hodis, Wayne Decatur