User:Laura Fountain/Chloride Ion Channel
From Proteopedia
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | {{STRUCTURE_1k0o | PDB=1k0o | SCENE=User:Laura_Fountain/Sandbox_1/1k0o/1}} | ||
== CLIC1: A Chloride Ion Channel == | == CLIC1: A Chloride Ion Channel == | ||
| - | + | The CLIC family consists of seven members: CLIC1-5, p64, and parchorin. CLIC1 is the most commonly studied member of the CLIC family because it is expressed to some extent in most tissues and cell types that have been studied and is particularly highly expressed in muscle.<ref name="Tulk">PMID:11940526</ref> Along with being present in the plasma membrane, CLIC1 has been found in various intracellular membranes, such as those of the mitochondria, nucleus (where it is designated NCC27), vesicles, and the endoplasmic reticulum.<ref name="Cromer">PMID:12202911</ref><ref name="Harrop">PMID:11551966</ref> | |
| - | + | This wide range of locations in the cell causes a plausible reason to assume that the CLIC chloride channel family participate in an equally wide variety of physiological processes. Some of these include cell division, kidney function, bone resorption, transepithelial transport, and signal transduction. <ref name="Cromer">PMID:12202911</ref> | |
| + | CLIC1 is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. When disrupted cells are washed approximately half of the CLIC1 proteins will remain within the fractioned membrane as would be expected from an integral membrane protein. Atypically, the other half will behave as a soluble cytoplasmic protein and exist within the aqueous extract.<ref name="Tulk">PMID:11940526</ref> Tulk et. al. showed that functionality of the protein isn't greatly effected by the method with which the protein inserts itself into the membrane. | ||
| - | + | This is part of the evidence which leads Tulk et. al. to postulate that CLIC1 is among the small group of proteins which are assembled as soluble cytoplasmic proteins, which will then insert themselves into the appropriate membrane via their own mechanism. | |
| - | + | == Structure == | |
| - | + | <applet load='1k0o' size='300' frame='true' align='right' caption='Dimer view of CLIC1' /> | |
| - | + | The CLIC family is defined by a COOH-terminal core segment of ~230 amino acids that are highly conserved among the family members. CLIC1 only contains a few amino acids upstream of the <scene name='User:Laura_Fountain/Sandbox_1/1k0o/3'>conserved core</scene>.<ref name="Tulk">PMID:11940526</ref> | |
| - | + | It has a homodimeric structure with one pore per subunit, which creates an incredibly unique "double barreled" channel. The <scene name='User:Laura_Fountain/Sandbox_1/1k0o/4'>N-domain</scene> of CLIC1 (~ amino acids 1-90) consists of <scene name='User:Laura_Fountain/Sandbox_1/1k0o/5'>4 beta-sheets</scene> and <scene name='User:Laura_Fountain/Sandbox_1/1k0o/6'>3 alpha-helices</scene>, and the <scene name='User:Laura_Fountain/Sandbox_1/1k0o/7'>C-domain</scene> consists entirely of alpha-helices. The <scene name='User:Laura_Fountain/Sandbox_1/1k0o/9'>long loop between helices</scene> at the foot of CLIC1 (Pro147–Gln164) is a distinctive feature of the CLICs. It is highly negatively charged with seven acidic residues.<ref name="Harrop">PMID:11551966</ref> | |
| + | |||
| + | Integration of CLIC1 into the membrane is a highly prospect mechanism, but it is likely to require a major structural rearrangement, probably of the <scene name='User:Laura_Fountain/Sandbox_1/1k0o/4'>N-domain</scene><ref name="Harrop">PMID:11551966</ref>, which would insert itself and then allow the <scene name='User:Laura_Fountain/Sandbox_1/1k0o/7'>C-domain</scene> helices to insert and form the pore. | ||
| + | |||
| + | == Selectivity for Chloride == | ||
| + | |||
| + | Based on the similarity of CLIC1's <scene name='User:Laura_Fountain/Sandbox_1/Channel/1'>antiparallel alpha helical loop</scene> (residues 101-145) to those of other better understood proteins which are able to insert themselves into the membrane, Tulk et. al. propose that this is the single area of the protein which is able to transverse the membrane and also serve as a pore. The <scene name='User:Laura_Fountain/Sandbox_1/Channel/3'>5 positively</scene> and <scene name='User:Laura_Fountain/Sandbox_1/Channel/4'>5 negatively</scene> charged amino acids on each end of the alpha helices could be part of the ion selectivity of this channel.<ref name="Tulk">PMID:11940526</ref> | ||
| + | |||
| + | == Potential Ion Gating == | ||
| + | |||
| + | At its binding site in the pore, chloride could interact with the ends of four helices that come from both sides of the membrane. A <scene name='User:Laura_Fountain/Sandbox_1/Channel/5'>glutamate residue</scene> that protrudes into the pore is proposed to participate in gating due to its negative charge.<ref name="CLC">PMID:12163078</ref> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| |||||||||
| 1k0o, resolution 1.75Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
CLIC1: A Chloride Ion Channel
The CLIC family consists of seven members: CLIC1-5, p64, and parchorin. CLIC1 is the most commonly studied member of the CLIC family because it is expressed to some extent in most tissues and cell types that have been studied and is particularly highly expressed in muscle.[1] Along with being present in the plasma membrane, CLIC1 has been found in various intracellular membranes, such as those of the mitochondria, nucleus (where it is designated NCC27), vesicles, and the endoplasmic reticulum.[2][3]
This wide range of locations in the cell causes a plausible reason to assume that the CLIC chloride channel family participate in an equally wide variety of physiological processes. Some of these include cell division, kidney function, bone resorption, transepithelial transport, and signal transduction. [2]
CLIC1 is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. When disrupted cells are washed approximately half of the CLIC1 proteins will remain within the fractioned membrane as would be expected from an integral membrane protein. Atypically, the other half will behave as a soluble cytoplasmic protein and exist within the aqueous extract.[1] Tulk et. al. showed that functionality of the protein isn't greatly effected by the method with which the protein inserts itself into the membrane.
This is part of the evidence which leads Tulk et. al. to postulate that CLIC1 is among the small group of proteins which are assembled as soluble cytoplasmic proteins, which will then insert themselves into the appropriate membrane via their own mechanism.
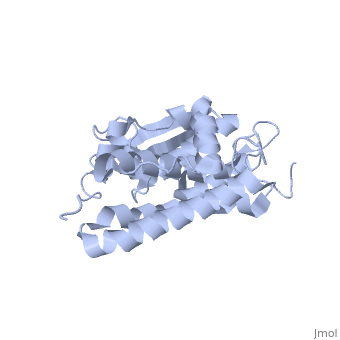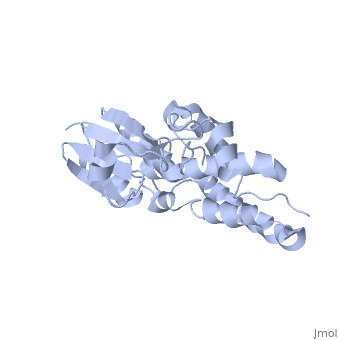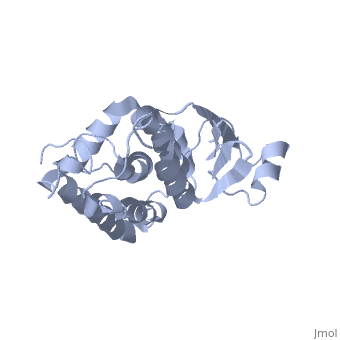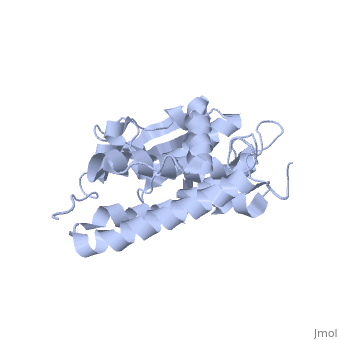
Structure
|
The CLIC family is defined by a COOH-terminal core segment of ~230 amino acids that are highly conserved among the family members. CLIC1 only contains a few amino acids upstream of the .[1]
It has a homodimeric structure with one pore per subunit, which creates an incredibly unique "double barreled" channel. The of CLIC1 (~ amino acids 1-90) consists of and , and the consists entirely of alpha-helices. The at the foot of CLIC1 (Pro147–Gln164) is a distinctive feature of the CLICs. It is highly negatively charged with seven acidic residues.[3]
Integration of CLIC1 into the membrane is a highly prospect mechanism, but it is likely to require a major structural rearrangement, probably of the [3], which would insert itself and then allow the helices to insert and form the pore.
Selectivity for Chloride
Based on the similarity of CLIC1's (residues 101-145) to those of other better understood proteins which are able to insert themselves into the membrane, Tulk et. al. propose that this is the single area of the protein which is able to transverse the membrane and also serve as a pore. The and charged amino acids on each end of the alpha helices could be part of the ion selectivity of this channel.[1]
Potential Ion Gating
At its binding site in the pore, chloride could interact with the ends of four helices that come from both sides of the membrane. A that protrudes into the pore is proposed to participate in gating due to its negative charge.[4]
References
- ↑ 1.0 1.1 1.2 1.3 Tulk BM, Kapadia S, Edwards JC. CLIC1 inserts from the aqueous phase into phospholipid membranes, where it functions as an anion channel. Am J Physiol Cell Physiol. 2002 May;282(5):C1103-12. PMID:11940526 doi:10.1152/ajpcell.00402.2001
- ↑ 2.0 2.1 Cromer BA, Morton CJ, Board PG, Parker MW. From glutathione transferase to pore in a CLIC. Eur Biophys J. 2002 Sep;31(5):356-64. Epub 2002 May 23. PMID:12202911 doi:10.1007/s00249-002-0219-1
- ↑ 3.0 3.1 3.2 Harrop SJ, DeMaere MZ, Fairlie WD, Reztsova T, Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Jankova L, Warton K, Bauskin AR, Wu WM, Pankhurst S, Campbell TJ, Breit SN, Curmi PM. Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution. J Biol Chem. 2001 Nov 30;276(48):44993-5000. Epub 2001 Sep 10. PMID:11551966 doi:10.1074/jbc.M107804200
- ↑ Estevez R, Jentsch TJ. CLC chloride channels: correlating structure with function. Curr Opin Struct Biol. 2002 Aug;12(4):531-9. PMID:12163078