Kwon sandbox
From Proteopedia
| (29 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<applet load='2A93-custom.pdb' size='400' frame='true' align='right' caption='c-Myc is a [http://en.wikipedia.org/wiki/DNA-binding_protein DNA binding protein]' scene='C-Myc/Custom/2'/> | <applet load='2A93-custom.pdb' size='400' frame='true' align='right' caption='c-Myc is a [http://en.wikipedia.org/wiki/DNA-binding_protein DNA binding protein]' scene='C-Myc/Custom/2'/> | ||
| - | + | ||
| + | == Introduction == | ||
| - | + | ---- | |
| - | + | <scene name='Kwon_sandbox/C-myc/1'>c-Myc</scene> is a protein that binds to DNA and regulates transcription, a transcription factor. Bishop and collegues discovered viruses that induced chicken sarcomas. They studied the virus and identified an oncogene that would cause uncontrolled cellular proliferation. The viral oncogene that caused the sarcomas was identified as v-myc. Later on the homologous gene in chickens was discovered, and called c-myc. These findings provided evidence that activated c-Myc proteins were significant in cellular growth regulation. | |
| - | + | ==Role in Cancer== | |
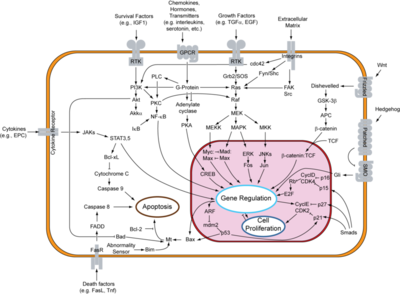
| + | [[Image:800px-Signal transduction v1.png|Apoptosis signal pathway|400 px|thumb]] | ||
| + | c-Myc is further along in the signal transduction pathway of the epithelial growth factor receptor (EGF receptor) which deals with the proliferation of cells. Mutations of c-Myc have a strong correlation to cancer. Normally c-myc is tightly regulated and c-Myc is short lived, but cancer cells express c-myc uncontrollably and are unable to degrade the c-Myc protein. This over expression and inability to rid the protein causes it to be active much longer. thus causing the over expression of genes needed for cell proliferation causing cancer. Over expression of c-Myc is prevalent in 80% of brest cancers, 70% colorectal cancers, 90% of gynecological cancers, 50% of hepatocellular carcinomas and is particularly prevalent Burkitt’s Lymphoma. | ||
| - | + | ==Research of Structure and Function== | |
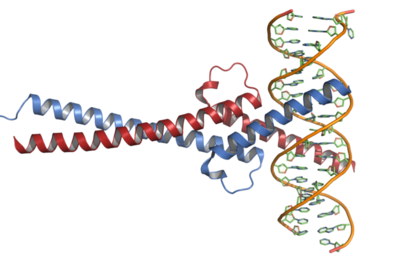
| + | [[Image:C-Myc-DNA complex.png|c-Myc Dna complex|400 px|thumb]] | ||
| + | There are two main structures of the c-Myc proteins that are significant in its function. These are the Thr58 sight and the helix-loop-helix (HLH) motif surrounded by a basic amino acid region and a leucine zipper motif. | ||
| - | == | + | Kandil and colleagues speculated that the carboxyl terminus of the c-Myc protein had a similar structure to that of the helix-loop-helix family of DNA-binding proteins. Their research showed that the <scene name='Kwon_sandbox/Hlh/1'>Helix-Loop-Helix Structure</scene> was in fact the <scene name='Kwon_sandbox/Dna_binding_domain/1'>DNA binding Domain</scene> of c-Myc and were able to establish the corresponding binding sequence as GACCACGTGGTC. This sequence was found to be present in regulatory regions of genes during replication. They compared DNA binding of c-Myc to HLH protein TFEB. They found that the two proteins had the same inner nucleotides, providing significant evidence of the homology. Kandil and colleagues then placed spacing between half-sites of the DNA binding site. The inability of c-Myc to bind to the altered site provided evidence that c-Myc dimerizes when bound to DNA. |
| - | Myc | + | Bahram and colleagues found that the mutation of <scene name='Kwon_sandbox/T58/1'>Thr58</scene> in c-Myc was prevalent in many cancers. They then researched the effect of Thr58 mutation and found that it was the ubiquitination site of the protein. Their in vitro experiment showed that c-Myc with Thr58 mutation had a longer turnover rate than wild type c-Myc. They also found that histadine-tagged ubiquitin octamers were unable to bind to Thr58 mutant c-Myc proteins but successfully did bind to wild type. This provided strong evidence that the site is indeed the ubiquitination site. |
| - | == | + | == References == |
| - | + | ---- | |
| + | Dang, C.V.(1999) c-Myc Target Genes Involved in Cell Growth, Apoptosis, and Metabolism. [http://www.ncbi.nlm.nih.gov/pubmed/9858526 Molecular and Cellular Biology], 19: 1-11. | ||
| - | + | Kandil, A.N. (1991) Determination of the c-MYC DNA-binding site. Proc. natl. Acad. Sci. USA Vol. 88, pp6162-6166, July 1991 Genetics | |
| - | + | Bahram et al., 2000; c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | Myc | + | |
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
|
Contents |
Introduction
is a protein that binds to DNA and regulates transcription, a transcription factor. Bishop and collegues discovered viruses that induced chicken sarcomas. They studied the virus and identified an oncogene that would cause uncontrolled cellular proliferation. The viral oncogene that caused the sarcomas was identified as v-myc. Later on the homologous gene in chickens was discovered, and called c-myc. These findings provided evidence that activated c-Myc proteins were significant in cellular growth regulation.
Role in Cancer
c-Myc is further along in the signal transduction pathway of the epithelial growth factor receptor (EGF receptor) which deals with the proliferation of cells. Mutations of c-Myc have a strong correlation to cancer. Normally c-myc is tightly regulated and c-Myc is short lived, but cancer cells express c-myc uncontrollably and are unable to degrade the c-Myc protein. This over expression and inability to rid the protein causes it to be active much longer. thus causing the over expression of genes needed for cell proliferation causing cancer. Over expression of c-Myc is prevalent in 80% of brest cancers, 70% colorectal cancers, 90% of gynecological cancers, 50% of hepatocellular carcinomas and is particularly prevalent Burkitt’s Lymphoma.
Research of Structure and Function
There are two main structures of the c-Myc proteins that are significant in its function. These are the Thr58 sight and the helix-loop-helix (HLH) motif surrounded by a basic amino acid region and a leucine zipper motif.
Kandil and colleagues speculated that the carboxyl terminus of the c-Myc protein had a similar structure to that of the helix-loop-helix family of DNA-binding proteins. Their research showed that the was in fact the of c-Myc and were able to establish the corresponding binding sequence as GACCACGTGGTC. This sequence was found to be present in regulatory regions of genes during replication. They compared DNA binding of c-Myc to HLH protein TFEB. They found that the two proteins had the same inner nucleotides, providing significant evidence of the homology. Kandil and colleagues then placed spacing between half-sites of the DNA binding site. The inability of c-Myc to bind to the altered site provided evidence that c-Myc dimerizes when bound to DNA.
Bahram and colleagues found that the mutation of in c-Myc was prevalent in many cancers. They then researched the effect of Thr58 mutation and found that it was the ubiquitination site of the protein. Their in vitro experiment showed that c-Myc with Thr58 mutation had a longer turnover rate than wild type c-Myc. They also found that histadine-tagged ubiquitin octamers were unable to bind to Thr58 mutant c-Myc proteins but successfully did bind to wild type. This provided strong evidence that the site is indeed the ubiquitination site.
References
Dang, C.V.(1999) c-Myc Target Genes Involved in Cell Growth, Apoptosis, and Metabolism. Molecular and Cellular Biology, 19: 1-11.
Kandil, A.N. (1991) Determination of the c-MYC DNA-binding site. Proc. natl. Acad. Sci. USA Vol. 88, pp6162-6166, July 1991 Genetics
Bahram et al., 2000; c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover