Urate Oxidase
From Proteopedia
(Difference between revisions)
| (51 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
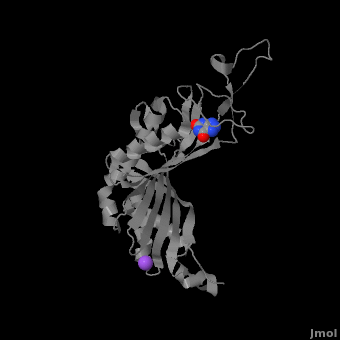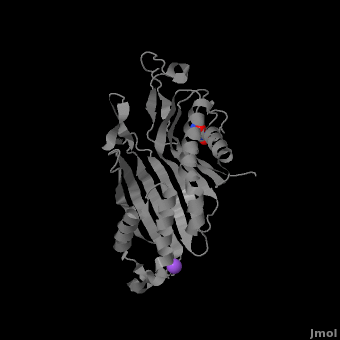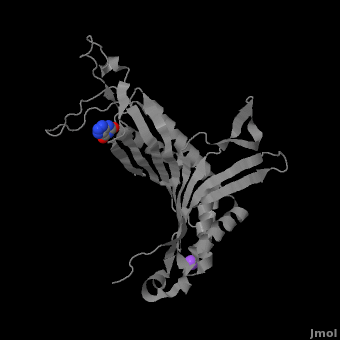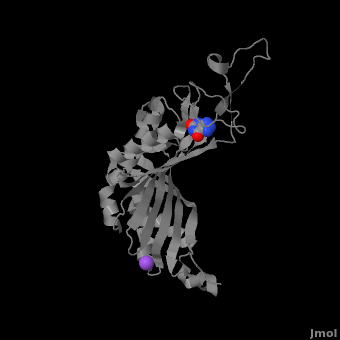
| - | + | <StructureSection load='' size='350' side='right' caption='Monomer of the tetrameric urate oxidase complex with 8-azaxanthine and Na+ ion (purple) [[3bk8]]' scene='Sandbox_185/Urate_oxidase_basic_monomer/1' > | |
| - | < | + | __TOC__ |
| - | + | ||
== Introduction == | == Introduction == | ||
| - | Urate oxidase | + | '''Urate oxidase''' or '''uricase''' or '''uric acid degradation bifunctional protein''' is an enzyme belonging to the purine degradation pathway, whose function is to prevent build-up of uric acid by catalyzing the oxidation of uric acid by molecular oxygen, leading to the production of 5-hydroxyisourate and hydrogen peroxide.<ref name="gabison">PMID:18638417</ref> The catalytic mechanism of urate oxidase is unique because it does not require a cofactor or metal ion.<ref name="gabison"/> This enzyme is found in many organisms, but not in higher apes and humans. It has been suggested that this may be an evolutionary advantage, as uric acid is a strong anti-oxidant.<ref name="gabison"/> The presence of uric acid therefore results in fewer free radicals and fewer instances of cancer as a result of aging.<ref name="gabison"/> The lack of urate oxidase does, however, result in elevated levels of uric acid in the plasma, which can be fatal. Urate oxidase was first isolated from the fungus ''Aspergillus flavus'', but is expressed in ''Saccharomyces cerevisiae'' for research and medical purposes.<ref name="gabison"/>. See also [[Uricase (Urate Oxidase)]]. |
== Structure == | == Structure == | ||
| Line 10: | Line 9: | ||
The complete functional structure of urate oxidase is a 135-kDa barrel-shaped homo-tetramer that has a height of 7 nm, with an inner radius of 0.6 nm and an outer radius of 3 nm.<ref name="colloch">PMID:9360612</ref><ref name="colloch2">Colloc'h N, Girard E, Dhaussy a, Kahn R, Ascone I, Mezouar M, Fourme R. High pressure macromolecular crystallography: the 140-MPa resolution of urate oxidase, a 135-kDa tetrameric assembly. Biochemica et Biophysica Acta - Proteins and Proteomics. 2006 March;1764:3.</ref> It has four identical active sites that are found at dimeric interfaces formed between the monomers and a central void tunnel of unknown function that is 5 nm long and has a diameter of 1.2 nm.<ref name="colloch"/> Each monomer, formed by 301 amino acid residues, has two structurally equivalent domains, which Colloc'h ''et al.'' termed “tunneling fold” domains (this feature allows urate oxidase to be placed in the expanding family of tunnel-shaped proteins that also includes 3i2b and 1a9c).<ref name="colloch"/> These domains are comprised of a four-strand long antiparallel beta sheet with two helices on the concave side. Together, the two tunneling fold domains of each monomer form an eight-strand long antiparallel beta sheet, where all four helices are found on the concave side of the sheet. The dimer forms an α<sub>8</sub>β<sub>16</sub> barrel, in which the eight helices make up the outer surface of the barrel.<ref name="colloch"/> | The complete functional structure of urate oxidase is a 135-kDa barrel-shaped homo-tetramer that has a height of 7 nm, with an inner radius of 0.6 nm and an outer radius of 3 nm.<ref name="colloch">PMID:9360612</ref><ref name="colloch2">Colloc'h N, Girard E, Dhaussy a, Kahn R, Ascone I, Mezouar M, Fourme R. High pressure macromolecular crystallography: the 140-MPa resolution of urate oxidase, a 135-kDa tetrameric assembly. Biochemica et Biophysica Acta - Proteins and Proteomics. 2006 March;1764:3.</ref> It has four identical active sites that are found at dimeric interfaces formed between the monomers and a central void tunnel of unknown function that is 5 nm long and has a diameter of 1.2 nm.<ref name="colloch"/> Each monomer, formed by 301 amino acid residues, has two structurally equivalent domains, which Colloc'h ''et al.'' termed “tunneling fold” domains (this feature allows urate oxidase to be placed in the expanding family of tunnel-shaped proteins that also includes 3i2b and 1a9c).<ref name="colloch"/> These domains are comprised of a four-strand long antiparallel beta sheet with two helices on the concave side. Together, the two tunneling fold domains of each monomer form an eight-strand long antiparallel beta sheet, where all four helices are found on the concave side of the sheet. The dimer forms an α<sub>8</sub>β<sub>16</sub> barrel, in which the eight helices make up the outer surface of the barrel.<ref name="colloch"/> | ||
| - | Within the active site, two residues, Arg 176 and Gln 228, are responsible for hydrogen-binding the substrate (uric acid).<ref name="gabison"/> Approximately 0.33 nm above the ligand, Asn 254 and Thr 57* (* indicates a residue from a different | + | Within the active site, two residues, <scene name='Sandbox_185/Urate_oxidase_basic_monomer/3'>Arg 176</scene> and <scene name='Sandbox_185/Urate_oxidase_basic_monomer/2'>Gln 228</scene>, are responsible for hydrogen-binding the substrate (uric acid).<ref name="gabison"/> Approximately 0.33 nm above the ligand, Asn 254 and Thr 57* (* indicates a residue from a different subunit), hydrogen-bond the molecular oxygen and the catalytic water molecule, depending on the step of the reaction.<ref name="gabison"/> The catalytic water molecule and the γ oxygen of Thr 57* are the beginning of a proton transfer chain that also involves Lys 10*, His 256 and two other water molceules, ending at N9 of uric acid.<ref name="gabison"/> |
| - | + | *<scene name='38/382957/Cv/3'>Whole active site</scene>. Water molecules are shown as red spheres. | |
== Function == | == Function == | ||
| - | Urate oxidase has the function of preventing uric acid levels from becoming too high in certain organisms, including bacteria, yeast, fungi and some mammals.<ref name="colloch2"/> In mammals, urate oxidase is found in the peroxisomes of liver hepatocytes. Although this enzyme has been found in many organisms, it is absent in higher apes and humans (in the human genome this is a result of a nonsense mutation in the codon for Arg 176).<ref | + | Urate oxidase has the function of preventing uric acid levels from becoming too high in certain organisms, including bacteria, yeast, fungi and some mammals.<ref name="colloch2"/> In mammals, urate oxidase is found in the peroxisomes of liver hepatocytes.<ref name="wu">PMID:1556746</ref> Although this enzyme has been found in many organisms, it is absent in higher apes and humans (in the human genome this is a result of a nonsense mutation in the codon for Arg 176).<ref name="wu"/> |
| - | Although the exact mechanism by which urate oxidase carries out its function is yet unknown, most studies agree that the catalytic proton transfer previously described is involved. After compiling data from multiple studies, Gabison ''et al.'' came up with a plausible mechanism. This mechanism involves sequential addition of molecular oxygen and a water molecule to the same catalytic site, known as the peroxo site (formed by Thr 57* | + | Although the exact mechanism by which urate oxidase carries out its function is yet unknown, most studies agree that the catalytic proton transfer previously described is involved.<ref name="gabison"/> After compiling data from multiple studies, Gabison ''et al.'' came up with a plausible mechanism. This mechanism involves sequential addition of molecular oxygen and a water molecule to the same catalytic site, known as the peroxo site (formed by the side chains of Asn 254 and Thr 57*), resulting in a mechanism with four intermediates leading up to the product 5-hydroxyisourate.<ref name="gabison"/> As other studies have previously proposed, Gabison ''et al.'' also suggest that after uric acid binds to the enzyme, a urate [N3<sup>-</sup>, N7<sup>-</sup>] dianion is formed.<ref name="gabison"/> This step is followed by the binding of molecular oxygen, leading to the production of a 5-hydroperoxyisourate intermediate.<ref name="gabison"/> Elimination of hydrogen peroxide from 5-hydroperoxyisourate then occurs, forming dehydrourate.<ref name="gabison"/> The final step of this reaction mechanism is the addition of a hydroxyl group derived from the catalytic water molecule to dehydrourate, followed by protonation, to give the product, 5-hydroxyisourate.<ref name="gabison"/> Protonation occurs via the proton transfer chain previously described.<ref name="gabison"/> |
| - | There is great interest in determining the exact mechanism of urate oxidase due to the fact that this enzyme does not require a cofactor or metal ion to carry out its function. | ||
== Medical Implications == | == Medical Implications == | ||
| - | |||
| - | Hyperuricemia is a condition in which uric acid levels in the blood become abnormally high. This may be caused by various factors including genetic disorders (Lesch-Nyhan syndrome), diet, kidney disease, and chemotherapy. Symptoms of hyperuricemia include joint inflammation (gout), kidney stones, pain and fever. | ||
In some patients with leukemia and lymphoma, chemotherapy causes the development of a condition known as tumor lysis syndrome (a complication of hyperuricemia, hyperphosphatemia and hyperkalemia).<ref name="renyi">PMID:17387390</ref> Tumor lysis syndrome results from the rapid breakdown of cells that occurs after chemotherapy, leading to significantly increased levels of uric acid and other compounds in the blood, eventually leading to kidney failure. | In some patients with leukemia and lymphoma, chemotherapy causes the development of a condition known as tumor lysis syndrome (a complication of hyperuricemia, hyperphosphatemia and hyperkalemia).<ref name="renyi">PMID:17387390</ref> Tumor lysis syndrome results from the rapid breakdown of cells that occurs after chemotherapy, leading to significantly increased levels of uric acid and other compounds in the blood, eventually leading to kidney failure. | ||
| - | Sanofi-Aventis, a European-based pharmaceutical company, produces a drug known as Fasturtec (rasburicase) that is a recombinant form of urate oxidase that is produced in a genetically modified strain of ''Saccharomyces cerevisiae''. This drug is administered to patients who are experiencing tumor lysis syndrome as a result of chemotherapy.<ref name="renyi"/> It is interesting to note that a non-recombinant form of urate oxidase, also produced by Sanofi Aventis and known as Uricozyme, has been used throughout some European countries for the same purpose, but it seems to cause an allergic reaction in 5% of patients, whereas the recombinant urate oxidase does not.<ref name="renyi"/> | + | Sanofi-Aventis, a European-based pharmaceutical company, produces a drug known as Fasturtec (rasburicase) that is a recombinant form of urate oxidase that is produced in a genetically modified strain of ''Saccharomyces cerevisiae''.<ref name="renyi"/> This drug is administered to patients who are experiencing tumor lysis syndrome as a result of chemotherapy.<ref name="renyi"/> It is interesting to note that a non-recombinant form of urate oxidase, also produced by Sanofi Aventis and known as Uricozyme, has been used throughout some European countries for the same purpose, but it seems to cause an allergic reaction in 5% of patients, whereas the recombinant urate oxidase does not.<ref name="renyi"/> |
| + | ==3D structures of urate oxidase== | ||
| + | [[Urate oxidase 3D structures]] | ||
| + | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | + | [[Category:Topic Page]] | |
| - | + | ||
Current revision
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Gabison L, Prange T, Colloc'h N, El Hajji M, Castro B, Chiadmi M. Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: mechanistic implications. BMC Struct Biol. 2008 Jul 20;8:32. PMID:18638417 doi:10.1186/1472-6807-8-32
- ↑ 2.0 2.1 2.2 2.3 Colloc'h N, el Hajji M, Bachet B, L'Hermite G, Schiltz M, Prange T, Castro B, Mornon JP. Crystal structure of the protein drug urate oxidase-inhibitor complex at 2.05 A resolution. Nat Struct Biol. 1997 Nov;4(11):947-52. PMID:9360612
- ↑ 3.0 3.1 Colloc'h N, Girard E, Dhaussy a, Kahn R, Ascone I, Mezouar M, Fourme R. High pressure macromolecular crystallography: the 140-MPa resolution of urate oxidase, a 135-kDa tetrameric assembly. Biochemica et Biophysica Acta - Proteins and Proteomics. 2006 March;1764:3.
- ↑ 4.0 4.1 Wu XW, Muzny DM, Lee CC, Caskey CT. Two independent mutational events in the loss of urate oxidase during hominoid evolution. J Mol Evol. 1992 Jan;34(1):78-84. PMID:1556746
- ↑ 5.0 5.1 5.2 5.3 Renyi I, Bardi E, Udvardi E, Kovacs G, Bartyik K, Kajtar P, Masat P, Nagy K, Galantai I, Kiss C. Prevention and treatment of hyperuricemia with rasburicase in children with leukemia and non-Hodgkin's lymphoma. Pathol Oncol Res. 2007;13(1):57-62. Epub 2007 Mar 27. PMID:17387390 doi:PAOR.2007.13.1.0057
Proteopedia Page Contributors and Editors (what is this?)
Sonja Senekovic, Michal Harel, Alexander Berchansky, David Canner, Andrea Gorrell, Joel L. Sussman